Repeated DNA sequences
advertisement

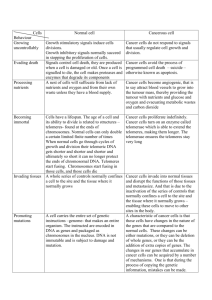
Repeated DNA sequences 4 Prof Duncan Shaw Molecular & Cell Biology Lecture 4 Telomeres - why they are necessary Structure of telomeres Synthesis of telomeres by telomerase Telomeres, cancer, and ageing Prolonging cellular lifespan Telomeres - why they are necessary There are 2 questions to solve concerning the ends of eukaryotic chromosomes: what stops them from being degraded by exonucleases or ligated to other free ends, and how is their length maintained? As the picture shows, the normal process of DNA replication would result in the end of the chromosome becoming shorter, since once the primer for DNA replication is removed, the singlestranded DNA opposite would be degraded. The answers came initially from molecular studies on the protozoan organism Tetrahymena. This is unusual in that it has a "macronucleus" in which there are many small chromosomal fragments, each carrying a few active genes and 2 telomeres. This is how the organism arranges its DNA for transcription, and it is useful for the study of telomeres because there is a very high ratio of telomeric DNA to total DNA. These "minichromosomes" can be isolated in a pure form. This showed that the telomeres consist of 20-70 tandemly repeated copies of the sequence ....CCCCAA..... The diagram shows an experiment to transfer Tetrahymena telomeres into yeast in order to study them further. The top frame shows the structure of the mini-chromosome, emphasising the telomeres. The repeats fold back at the end so that singlestranded ends are not left dangling, but the structure is not completely ligated together since there are nicks on one strand. In the yeast experiment this DNA is digested and ligated to a marker gene selectable in yeast (leu2). After transformation and selection the DNA forms stable linear "plasmids" which can be isolated and sequenced. However when these plasmids had been grown in yeast for several generations, then isolated, it was found that the sequence of some of the telomeric repeats had changed to ...CnA.... where n is 1-3. So the yeast adds different telomeric sequences, and it can't be using the Tetrahymena telomeres as a template (in contrast to normal DNA replication). The yeast plasmid can also be used as a vector to clone other telomeres, by digesting it with a restriction enzyme, ligating one half of it to a digest of the target DNA, and selecting clones that are stably maintained in yeast. Since 2 telomeres are needed for stability, any clones like this must have acquired their 2nd telomere from the target DNA. Telomeres seem to be functional between species so this experiment can be used to isolate human telomeres, for example. Further studies have shown that all known eukaryotic telomeres have similar structures, with short C+Arich repeats. A culture of Tetrahymena be synchronised so that they are all at the stage in the cell cycle. By analysing the DNA at various timepoints, the synthesis of telomeres can be followed by use of Southern blots and a telomere repeat probe (see diagram). This shows that the rate of telomere addition is about 7-10bp per cell division, sufficient to maintain the telomeres at a constant size over longer periods. Another observation from this experiment is that there is heterogeneity in telomere length, because not exactly the same number of repeats is added to each telomere at each cell division. This is seen as a smear of hybridising DNA on the Southern blot, rather than a single band. Telomeres are built by an enzyme, telomerase. It adds copies of the telomeric repeat sequence (TTGGGG for Tetrahymena) to the end of the chromosome. To do this it has an internal RNA template with the sequence complementary to the repeat, ....CAACCCCAA....Each species' telomerase carries an RNA template with the sequence that determines the telomere sequence for the species. This explains why yeast add yeast telomere repeats to the Tetrahymena telomeres in the experiment above. This picture shows the mechanism of telomere synthesis. The loop structure at the end is stabilised by base pairing of a non-standard type between the G bases. This has to unfold to allow telomerase to add new repeats, after which it refolds, DNA polymerase fills in the gapped region, and there is incomplete ligation leaving a few nicks. The nicks allow the DNA to unfold again at the next round of replication. Although this mechanism was worked out by studies on micro-organisms, it appears to be generally true for all eukaryotes. Telomeres, ageing, and cancer References for this section: Harley et al (1994). Cold Spring Harbor Symp. Quant. Biol. vol LIX, pp307-315 Bodnar et al (1998). Science 279, 349-352 Although single-celled organisms must maintain their telomeres so that when they divide they pass them on intact to their daughter cells, the same is not true of somatic tissues in multi-cellular organisms, which only have to divide for a limited number of times before the tissue they are in is fully developed. Therefore, their telomeres do not have to be maintained at the same length. Of course, germline cells do have to maintain their telomeres since they must go on dividing indefinitely. Somatic cells that are grown in culture will stop growing after 50-100 generations (senescence). This figure is reduced for cells derived from an elderly donor. Thus, they appear to have a "mitotic clock" that tells them when they are old, and so to stop dividing. In the body this mechanism would guard against proliferation of very old cells, that might have acquired harmful mutations over the course of their lifetime. In fact this mechanism is thought to guard against cancer. The diagram shows the multi-stage progression of a typical tumour. As the probability of the genetic mutation at each stage is low (about 1 in a million) it could take at least 80 cell divisions for the original cell to become a tumour, longer than the normal lifespan of a somatic cell. To study what is happening to telomeres in normal cell growth and in cancer, you need to measure their length. This is easily done using a Southern blot probed with the telomere repeat, as shown. It normally gives rise to a smear of DNA rather than a single band because within a population of cells there is variation in the number of repeats at each telomere. What we are interested in, is the average length and how it changes with time. In the diagram it is shown as decreasing. To test the idea that telomeres may be maintained in tumours, so that they carry on dividing (i.e. they have gained "immortality"), telomere length and telomerase enzyme activity were measured in a number of cell types, as shown in this table: Cell type Telomere length Telomerase activity Cultured "mortal" shortens 0 Cultured "immortal" stable + Normal testis stable + Normal somatic shortens 0 Tumours (breast, ovary, colon, stable or shortens + in most cases prostate etc) Telomerase is more likely to be active in late-stage tumours, and may indicate a poor prognosis. Thus to summarise: Telomerase is required to be active in single-cell organisms, and in germline cells of complex organisms, since these need to divide indefinitely Telomerase is not required in somatic cells, which have a finite lifespan; this is a built-in mechanism to limit the life of cells that might carry harmful mutations The development of a tumour may require telomerase to be activated so that the tumour cells can become immortal The picture shows the time course of telomere length in various conditions. In the top frame, germline cells (red line) maintain their telomeres. Somatic cell telomeres (blue) normally decline steadily till they reach a limit (M1, named "Hayflick limit" after its discoverer) where some telomeres are getting too short for comfort. Activation of an oncogene (black) may allow cells to ignore this "warning". A second limit (M2) where most telomeres have got extremely short is the signal for cells to undergo apoptosis (programmed cell death). In a tumour where telomerase has re-activated (red) this limit can be ignored and cells with short telomeres can carry on dividing. It has been suggested that inhibition of telomerase could be a good way to inhibit the growth of tumours without affecting normal somatic tissues. The second frame in the picture above shows the predicted effects of this (purple arrows). Tumour cells might be forced to undergo apoptosis when they reach the M2 limit. This treatment, if it were to become reality, might cause problems for reproduction since germline cells also need telomerase. Prolonging cellular lifespan If it is true that telomere length is the mitiotic clock that tells cells when they are too old to divide, then it should be possible to test this by activating telomerase in a cell line and causing it to go on dividing for ever. Bodnar et al did this by transfecting 2 human cell lines, retinal pigment epithelium and fibroblast, with a vector that expresses telomerase (top frame). They checked that this activated telomere lengthening with the Southern blot assay (middle frame). They then grew both cell lines, together with untransfected controls, in cell culture. The lifespan of the telomerase-actice cells was prolonged by up to 35 generations (bottom frame), and at the time their paper was published, they were still going strong. Apparently this experiment caused no ill-effects to the cells (i.e. they were phenotypically and karyotypically normal) but who knows what would happen in the long term? Cells with active telomerase do not necessarily become oncogenic (e.g. germline tissue), but it is reasonable to assume that the more times a cell divides, the more likely it is to gain harmful mutations. It is also necessary to control the activity of telomerase to achieve a balance between it and the tendency of telomeres to shorten. The authors of the study suggest some uses for cell lines immortalised by telomerase, including in research (to replace tumour cell lines by something more "normal" for cell biology and biochemistry studies), in biotechnology (production of useful molecules in culture) and in medicine (cellular or gene therapy).