Word file
advertisement

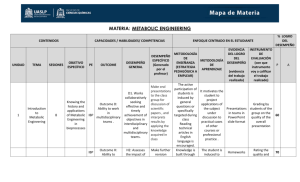
Supplementary Figure Legends Supplementary Figure 1. Id2 is degraded by APC/CCdh1 and binds to the core subunits of APC/C and Cdh1. a, Proteins associated with Id2 in neuroblastoma cells. Cellular extracts prepared from IMR-32 stably expressing Flag-HA-Id2 (FHId2) or the empty vector (Vec) were sequentially immunoprecipitated with Flag and HA antibodies. Associated proteins were analyzed by Silver stain. The indicated proteins were identified by LC-MS/MS. b, Endogenous Id2 protein is destabilized by Cdh1. U2OS cells were transfected with a Cdh1 expression vector or the empty vector and analyzed after 12 h by Western blot (shown in Fig. 1c). Parallel cultures were assayed by Flow cytometry. c, Immunoblot analysis of U2OS cells that were engineered to produce Cdh1 after removal of tetracycline (Tet). Where indicated the proteasomal inhibitor LLNL was added. d, Lysates from cells treated with tet or released from tet for 12 h were immunoprecipitated with E47 antibody and analyzed by Western blot for Id2 (E47-bound Id2). Total Id2, Cdh1 and actin are shown. e, Hela cells were transfected with expression plasmids for Id2 (wild type Id2 or 15 amino acid Nterminal deletion mutant Id2) and E47 in the presence or absence of Cdh1 and analyzed by Western blot. f, Interaction between Id2 and Cdh1. Hela cells were transfected with the indicated expression plasmids and left untreated or treated with MG-132 for 6 h before lysis. Lysates were reciprocally immunoprecipitated with Flag and HA antibodies followed by Western blot with antibodies against HA and Flag, respectively. g, Endogenous APC/C subunits and Cdh1 activator are associated with Id2 in neuroblastoma cells. IMR-32 cells were treated with 1 MG132 before lysis. Lysates were immunoprecipitated with rabbit IgG or a polyclonal antibody against Id2 and analyzed by Western blot for the indicated APC/C components. Input, 0.5% of total extract used for each immunoprecipitation. Supplementary Figure 2. Id2 is unstable in cells withdrawing the cell cycle. a, Immunoblot of Id2 in NIH3T3 cells deprived of serum for 12 h. b, CHX was added and extracts were prepared at the indicated times and analyzed by immunoblot. Double amount of total lysates was loaded for the serum deprived set and the exposures of the Id2 blots were normalized to show comparable levels of Id2 at the zero time point. Supplementary Figure 3. Id2 protein stability in quiescent cells is regulated by Cdh1. a, U2OS-Id2 cells in exponential phase of growth (0) were deprived of serum for the indicated times in the absence (-) or in the presence (+) of MG-132 for 6 h and analyzed by immunoblot. b, Pulse-chase analysis of U2OS cells stably expressing Id2 in the presence of serum (+ FBS) or after 48 h of serum deprivation (- FBS). Id2 was recovered by anti-Id2 immunoprecipitation and detected by autoradiography. c, Immunoblot of Id2 in SK-N-SH human neuroblastoma cells treated with vehicle (Exp) or RA for 48 h before addition of CHX for the indicated times (bottom panels). Cell extracts were analyzed by immunoblot. The exposures of the Id2 blots were normalized to show comparable levels of Id2 at the zero time point. d, U2OS-Id2 cells were 2 transfected with siRNA corresponding to Lamin, firefly luciferase (CTR), or Cdh1 mRNA and analyzed 16 h after transfection. Exogenous Id2 and endogenous Id1 were detected by immunoblot. e, U2OS-Id2 cells were cultured at low density and after transfection with CTR or Cdh1 siRNA were grown in the presence of serum or starved from serum for the indicated times. f, Quantification of band intensities from Fig. 1g, showing the turnover of endogenous Id2 of quiescent LAN-1 neuroblastoma cells in the presence and absence of Cdh1. The Id2 halflife is 3.7 minutes in control-treated cells and 36.6 minutes in Cdh1-knocked down cells. g, Immunoblot of Hela cells transfected with Id2 in the presence or absence of an Emi1 expression vector. Supplementary Figure 4. Degradation of Id protein by APC/CCdh1 is dependent on D box. a, Hela cells were transfected with wild type Id2 (Id2) or Id2-DB carrying deletion of amino acids 100-107 in the presence or absence of Cdh1 and analyzed by immunoblot. Hela cells were transfected with Id2HLH (b), Id1 (c), Id3 (d) or Id4 (e) with or without Cdh1 and analyzed by immunoblot. Supplementary Figure 5. Dual interaction modules of Id2 with Cdh1 and core APC/C. a, Immunoblot for Cdh1, Cdc27, Apc1 and actin from binding assay using GST, GST-Id2, GST-Id2-DBM and GST-Id2DB and extracts from Hela cells. b, Hela cells were transfected with Flag-Id2 or Flag-Id2-DBM expression plasmids. Lysates were immunoprecipitated with Flag antibody followed by Western blot. Input, 0.5% of total extract used for each immunoprecipitation. c, 3 Immunoblot analysis of U2OS cells stably expressing Id2 or Id2-DBM after treatment with TGF for 48 h. d, Autoradiogram of 35S-labelled Id2 after in vitro ubiquitination by immunopurified APC/C in the presence or in the absence of in vitro translated Cdh1 at the indicated times. Arrowhead indicates unmodified Id2. Supplementary Figure 6. Id2 does not affect the integrity and activity of APC/CCdh1 a, IMR-32 cells were infected with Ad-vect or Ad-Id2 and cell extracts were immunoprecipitated with Cdc27 antibody or mouse IgG followed by immunoblot for the indicated proteins. Input is 0.5% of the lysate used in the immunoprecipitation. b, Immunoblot of APC/CCdh1 targets from IMR-32 cells infected with Ad-vect or Ad-Id2. c, Autoradiogram of 35S-labelled cyclin B after in vitro degradation assay by immunopurified APC/C in the presence (+) or in the absence (-) of in vitro translated Cdh1 at the indicated times. Supplementary Fig. 7. Id2 is a target of APC/CCdh1 for axonal growth. a, Id2 associates with APC/C core subunits in the brain. Extracts from mouse brain at E16 were immunoprecipitated with Id2 antibody or rabbit IgG followed by immunoblot. b, Cerebellar granule neurons express Id2 during in vitro axonal elongation (1 to 4 days). 293T cells treated with CTR (-) or Id2 (+) siRNA are shown as a control for Id2 immunoblot. c, Expression of Id2 in granule neurons is markedly increased by proteasomal inhibition with MG-132. 293T are as in b. d, Phase contrast microscopy of neuronally differentiated SK-N-SH after sequential tretment with RA (5 days) and BDNF (4 days). e, Id2 associates with Cdh1 and 4 the core APC/C in terminally differentiated SK-N-SH. RA/BDNF treated SK-N-SH were exposed to MG132 before lysis. Lysates were immunoprecipitated with rabbit IgG or polyclonal anti-Id2 antibody and analyzed by Western blot for the indicated APC/C components. f, Silencing of Cdh1 in neuronally differentiated SK-N-SH stabilizes Id2 and Id1. RA/BDNF treated SK-N-SH were transfected with siRNA corresponding to firefly luciferase (CTR) or Cdh1 mRNA and analyzed 16 hours after transfection by Western blot for the indicated proteins. g, Cerebellar granule neurons were transfected 24 h after plating with CTR or Cdh1 siRNA and analyzed by immunoblot (shown in Fig. 3c). Parallel culture were immunostained for BrdU (top panels) and nuclei were counterstained with DAPI (middle panels). Arrowheads indicate a BrdU positive glial cell as shown in the bright field microphotograph in the bottom panels. h, Cerebellar granule neurons were transfected with vector or Id2-DBM and GFP expression plasmids and immunostained 2 days later with antibodies against GFP and MAP-2. MAP-2 staining is excluded from the long axonal projections (arrowheads, magnification 40X). Supplementary Fig. 8. Degradation resistant Id2 promotes axonal growth of cerebellar granule neurons in vitro and in vivo. a, Cerebellar granule neurons were transfected with vector, Id2 or Id2-DBM and GFP expression plasmids. Neurons were then grown in serum free medium and 3 days later were immunostained with an antibody against GFP. Asterisks and arrowheads indicate the cell body and axons, respectively. Scale bars equal to 50 m. b, Axonal 5 length was measured in granule neurons at DIV3. At least 600 cells were analyzed for each transfection. Values represent mean±SEM of triplicate experiments (*, p=<0.01). c, Axonal length was measured in cerebellar granule neurons in vivo. Neurons from P6 rat pups were transfected in suspension with the indicated plasmids and a GFP expression construct and plated on top of P9 cerebellar slices. Slices were analyzed 72 h later by immunohistochemistry for GFP antibody to label transfected cells and Hoechst to reveal the anatomical organization of cerebellar cortex. WM, white matter; IGL, internal granule layer. d, Axonal length was measured in at least 150 neurons for each construct from the experiments shown in panel c. Values represent mean±SEM of duplicate experiments (*, p= <0.0001). Supplementary Figure 9. A functional Cdh1-Id-bHLH pathway controls axonal growth. a, Induction of axonal growth inhibitory genes by E47 in neuroblastoma cells. SK-N-SH cells were infected with Ad-GFP or Ad-E47 and analyzed by microarray 8 h and 20 h after infection. Analysis at 20 h is from two independent experiments. b, RNA expression levels of E47 target genes was determined by qRT-PCR in SK-N-SH harvested 20 h after infection with Ad-GFP and 8 h and 20 h after infection with Ad-E47. Bars represent the mean and SD of triplicate experiments. Values are the fold changes above the Ad-GFP. c, Transcription from an E-box-luciferase plasmid was measured after transfection of E47 and increasing amounts of U6shCdh1 in SK-N-SH. Luciferase values were normalized towards co-transfected galactosidase. d, RNA expression levels for 6 the indicated genes were determined by qRT-PCR in SF210 harvested 24 hours after transfection with control siRNA or siRNA targeting Cdh1. Values represent the mRNA of Cdh1 siRNA transfectants expressed as percentage of the corresponding control. e, RNA expression levels for the indicated genes were determined by qRT-PCR in SK-N-SH transfected with control siRNA or siRNA targeting Cdh1 and then infected with Ad-GFP or Ad-E47. Values are plotted as fold induction by Ad-E47 over Ad-GFP. Supplementary Figure 10. Silencing of Cdh1 in cortical neurons stabilizes Id2 and Id1. DIV7 cortical neurons were transfected with siRNA corresponding to firefly luciferase (CTR) or Cdh1 mRNA and analyzed by Western blot for the indicated proteins. 7