Identification and functional characterization of a novel mutation in
advertisement

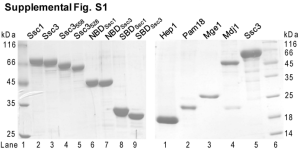
SUPPORTING INFORMATION SUPPLEMENTAL MATERIAL AND METHODS Functional complementation assay of Ssc1, yeast counterpart of mortalin Plasmids containing a TRP1 marker encoding wt Ssc1 or its variants (R103W, A453T, P486S) were transformed into a yeast strain Ssc1-wt, in which chromosomal Ssc1 (human mortalin homologue) is deleted and wt Ssc1 is expressed with a plasmid carrying URA3 marker. After transformation, drop dilution assays were performed on YPD plates as well as SD-Trp plates containing 5-FOA. The Latter allowed counterselection against cells containing the plasmid with the URA3 marker, as a toxic 5-fluorouracil was generated. SUPPLEMENTAL FIGURES Figure S1 Efficiency of mortalin knockdown and resulting reduction in mitochondrial mass by measuring TOM20 protein level in SH-SY5Y cells (A) Compared with untreated SH-SY5Y cells and control siRNA treated cells, the treatment with mortalin siRNA resulted in a substantial decrease in mortalin expression in SH-SY5Y cells compared to controls (data not shown). Densitometric analyses of the Western blot studies revealed a knockdown efficiency of more than 70% in SH-SY5Y cells treated with mortalin siRNA compared to control siRNA-treated cells. (B) Western blot analysis of SH-SY5Y cells treated with mortalin siRNA or control siRNA showed reduced protein levels of the mitochondrial marker Tom20 (data not shown). Densitometric analysis of average Tom20 levels confirmed the observed reduction in mitochondrial mass. TOM20 levels were normalized to β-actin levels, n=3. Figure S2 2 Analysis of LC3-I and LC3-II levels levels in control miRNA-treated SH-SY5Y cells Immunoblotting analysis of LC3 in SH-SY5Y cells stably expressing control miRNA treated with control medium conditions (CM) or starvation (EBSS) in the absence or presence of BafA1. Standard anti-LC3 ECL was conducted and tubulin used as loading control. Figure S3 Analysis of total endogenous WIPI-2 area in mortalin knockdown SH-SY5Y cells ImageJ-based analysis of the total WIPI-2 fluorescent area in both presence or absence of mortalin as described in Figure 4C. Figure S4 Analysis of endogenous p62 inclusions per cell in mortalin knockdown SH-SY5Y cells Automated high-throughput image acquisition and analysis was employed as previously described37 to measure endogenous p62 inclusions per cell in SH-SY5Y cells stably expressing mortalin miRNA or control miRNA under control medium conditions (CM) or starvation (EBSS) in the absence or presence of BafA1. Analysed cell counts are given below the statistics for each condition. Figure S5 Induction of autophagy results in decreased apoptotic cells death in mortalin knockdown cells SH-SY5Y cells were treated with control siRNA or mortalin siRNA and analyzed for apoptotic cell death via Annexin V staining measured by FACS analysis. Knockdown of mortalin resulted in an increased Annexin V signal. Treatment with Rapamycin led to a full rescue of this phenotype. 3 Figure S6 Functional complementation assay in yeast (A) Cells harboring plasmid-encoded Ssc1 variants were tested by dilution in 10-fold increments for their ability to grow at the indicated temperature on glucose-containing rich medium (Yeast Extract Peptone Dextrose YPD; left panel) or on synthetic medium lacking tryptophan and supplemented with 5-FOA (5-FOA SD-Trp; right panel). The latter condition favour the loss of a URA-marker containing plasmid that encodes wt Ssc1 since the URA3 gene product converts the nontoxic 5-FOA compound to toxic 5-fluorouracil. Hence, only cells expressing functional Ssc1 from plasmids containing the TRP1 marker were able to grow, which allowed us to assess to what degree PD-associated variants (R103W Ssc1 ≙human R126W mortalin; P486S Ssc1 ≙ human P509S mortalin) impair Ssc1 function. We confirmed a severe growth defect for cells harbouring the ATPase domain mutant R103W Ssc1. Cells complemented with P486S or A453T Ssc1 showed no growth defect suggesting that these mutations do not affect the function of Ssc1 yeast. Our findings support the notion of differential effects of mtHsp70 variants located in the ATPase domain or in the substrate binding domain in yeast. (B) Immunoblotting was performed to control for the expression of R103W Ssc1 on plasmid with TRP1 marker. Cells depleted of chromosomal Ssc1 and expressing wt Ssc1 from an URA3 marker plasmid (6xHis tag) were transformed with an empty TRP1 marker vector (vector, lane 1) or with a TRP1 marker vector encoding either wt SSC1 (lane 2) or the R103W mutant (lane 3). Whole cell extract was analyzed by SDS-PAGE and immunostaining with an antibody against Ssc1. Bands representing wt Ssc1 with a His-tag (URA plasmid) or the Ssc1 variants from the Trp plasmid are indicated.