tpj12593-sup-0008-MethodsS1
advertisement

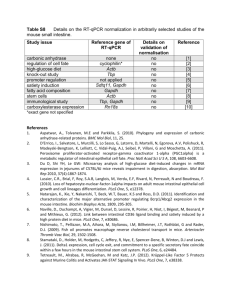
Methods S1. Supplementary experimental procedures. RT-qPCR analysis for detection of Pm8 and Pm3 expression Expression of Pm8 and Pm3 was quantified in a reverse transcription, quantitative realtime polymerase chain reaction (RT-qPCR) assay using a CFX96 Real-Time System C1000TM Thermal cycler (Bio-Rad, http://www.bio-rad.com/) and for the Chinese Spring x Kavkaz/4*Federation cross an Applied Biosystems 7500 Fast Real-Time PCR System (Life Technologies, http://www.lifetechnologies.com/). The translocation lines (Figure 4a) and the lines of the cross Chinese Spring x Kavkaz/4*Federation (Figure S3) were grown at 17°C, 70% relative humidity and 16 h light. Ten-day-old plants were then either infected with the Pm8 avirulent powdery mildew isolate 07230 or left without infection and covered with gas permeable covers. For the translocation lines leaf samples were taken after eight hours from infected and non infected plants and after 24 hours from infected plants. For lines of cross Chinese Spring x Kavkaz/4*Federation leaf samples were taken after 24 hours from infected and non-infected plants. Leaf samples were taken from ten-day-old transgenic plants (Figure 4b) which were grown at cycles of 16 h at 20°C with light and 8 h at 16°C in the dark at a constant relative humidity of 70%. RNA extraction and first-strand cDNA synthesis of 1 μg RNA were performed as described by Hurni et al. (2013). RT-qPCR primers and probes for the targets Pm8 and Pm3 and the reference genes were adopted from earlier studies (Travella et al., 2006; Brunner et al., 2011; Hurni et al., 2013) and are described in Tables S3 and S4. For the translocation lines and transgenic crosses (Figure 4), RT-qPCR was performed with 4 µl of tenfold diluted cDNA, forward and reverse primers according to Table S3 and 5 µl of KAPA PROBE FAST qPCR Master Mix (KK4701; Kapa Biosystems, http://www.kapabiosystems.com/) for Pm8 and Pm3 and KAPA SYBR® FAST qPCR Master Mix (KK4601; Kapa Biosystems) for ADP and TA.6863 in a total reaction 1 volume of 10 µl. Thermocycling conditions were 95°C for 20 s, followed by 40 cyles of 95°C for 3 s, then 60°C for 20 s. Specificities of the amplicons were checked by examination of dissociation curves with CFX Manager 3.1 Software (Bio-Rad) for the two SYBR® green-based targets ADP and TA.6863. RT-minus controls were checked with the reference gene target ADP. All RT-minus controls had quantification cycle (Cq) values of at least 12 cycles above the corresponding RT-plus samples, thus DNA contamination was irrelevant (>0.025%). The primer-probe combination for Pm8 showed no fluorophor signal on cDNA of lines homozygous for Pm3 and vice versa, therefore RT-qPCR assay specificities for the orthologous genes Pm8 and Pm3 were confirmed. Sensitivities of Pm8 and Pm3 qPCR assays were compared on the basis of plasmid DNA containing either the cloned Pm8 or the Pm3 RT-qPCR amplicon. At identical plasmid concentrations equal Cq values were obtained when the same threshold values were applied for both targets, hence both assays had the same sensitivity and expression levels were directly comparable among Pm8 and Pm3. Relative quantities were calculated and normalized to the two reference genes ADP and TA.6863 (CNRQ) using the program qbase+ V 2.6 (Biogazelle, http://www.biogazelle.com/). Target-specific amplification efficiencies are given in Table S3. For a description of efficiency calculation and RT-qPCR set up see Risk et al. (2012). RT-qPCR was performed with the lines from the cross Chinese Spring x Kavkaz/4*Federation with 4 µl of tenfold diluted cDNA, forward and reverse primers according to Tables S4 and 6 μl of TaqMan® Fast Universal PCR Master Mix (4352046; Life Technologies) for Pm8 and Pm3 and CDCP Fast SYBR® Green Master Mix (4385612; Life Technologies) for GAPDH in total reaction volumes of 16 µl. Thermocycling conditions were 95°C for 20 s, followed by 40 cyles of 95°C for 4 s, then 60°C for 30 s. For the SYBR® green based target GAPDH specificity of the amplicon 2 was checked by examination of dissociation curves with 7500 Software v2.0.6 (Life Technologies). RT-minus controls were checked with the target GAPDH. All RT-minus controls had quantification cycle (Cq) values of at least 12 cycles above the corresponding RT-plus samples, thus DNA contamination was irrelevant (>0.025%). Relative quantities were calculated and normalized to the reference genes GAPDH and CDCP (CNRQ) using the program qbase+ V 2.6 (Biogazelle). Target-specific amplification efficiencies are given in Table S4. Data analysis was performed using the statistical package JMP version 9.0 (SAS Institute, http://www.sas.com/). Tukey’s honestly significant difference test for the transgenic lines (Figure 4b) was done on log10 transformed expression values. In Figure 4b, the untransformed means are given and back-transformed 95% confidence intervals are indicated. REFERENCES Brunner, S., Hurni, S., Herren, G., Kalinina, O., von Burg, S., Zeller, S.L., Schmid, B., Winzeler, M. and Keller, B. (2011) Transgenic Pm3b wheat lines show resistance to powdery mildew in the field. Plant Biotechnology Journal, 9, 897910. Hurni, S., Brunner, S., Buchmann, G., Herren, G., Jordan, T., Krukowski, P., Wicker, T., Yahiaoui, N., Mago, R. and Keller, B. (2013) Rye Pm8 and wheat Pm3 are orthologous genes and show evolutionary conservation of resistance function against powdery mildew. The Plant Journal, 76, 957-969. Risk, J.M., Selter, L.L., Krattinger, S.G., Viccars, L.A., Richardson, T.M., Buesing, G., Herren, G., Lagudah, E.S. and Keller, B. (2012) Functional variability of the Lr34 durable resistance gene in transgenic wheat. Plant Biotechnology Journal, 10, 477-487. Travella, S., Klimm, T.E. and Keller, B. (2006) RNA interference-based gene silencing as an efficient tool for functional genomics in hexaploid bread wheat. Plant Physiology, 142, 6-20. 3