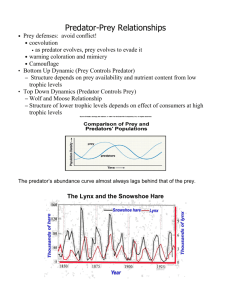
Author template for journal articles
advertisement

Table A1. Summary of the variables used/calculated by the model. User: Parameters set by the user before running the simulations. Model Structure: These variables are set automatically when the initial species abundances are generated according to the model structure. Simulation: These variables change constantly during the simulation. Variable Number of time steps Initial number of species Final number of species Initial number of basal species Number of top species Number of intermediate species Number of basal species Number of herbivores species Number of carnivore species Number of omnivore species Number of species presenting cannibalism Fraction of top species Fraction of intermediate species Fraction of basal species Fraction of herbivore species Fraction of omnivore species Fraction of carnivore species Fraction of caníbal species Initial Mean Trophic Level mean trophic level Standard Deviation of trophic level maximum trophic level Links between top and intermediate species Links between top and basal species Links between two intermediate species Links between intermediate and basal species links per species Links Standard Deviation of linkedness Initial Connectance Final connectance clustering coefficient Standard Deviation of generality Standard Deviation of vulerability mean chain length Standard Deviation of chain length maximum chain length Type of Functional Response Predator Interference Carrying capacity Metabolic type size ratio mean Amplitude mean Biomass mean Standard Deviation mean Variance mean CoVariance Abbreviation Type t S0 S B0 T I B Herb Carn Omn Can %T %I %B %Herb %Omn %Carn %Can TL0 TL TL.SD TLmax links ti links tb links ii links ib L/S L LinkSD C0 C Cl GenSD VulSD ChLen ChSD ChMax F c K User User Simulation User Model Structure Model Structure User Model Structure Model Structure Model Structure Model Structure Model Structure Model Structure User Model Structure Model Structure Model Structure Model Structure Model Structure Simulation Simulation Simulation Model Structure Model Structure Model Structure Model Structure Model Structure Simulation Simulation User Simulation Simulation Simulation Simulation Simulation Simulation Simulation User User User User User Simulation Simulation Simulation Simulation Simulation Ampli Biom StnDv Var CoVar 1 Table A2. Pearson correlation and p-value when comparing the response variables (Normalized Biomass Size-Spectra slope = NBSS slope, and the Pareto Shape Parameter = Pareto’s ) against the predictor variables not considered in main manuscript. n = 10000 Variable S T I B Herb Omn Can %T %I %B %Herb %Omn %Can TL TL.SD TLmax links ti links tb links ii links ib L/S L LinkSD Cl GenSD VulSD ChLen ChSD F Ampli Biom StnDv Var CoVar NBSS slope Correlation p-value -0,2861452 3,20E-121 0,07304625 4,66E-09 -0,2367817 1,67E-82 0,00922782 0,45979028 0,02882955 0,02089839 -0,1624816 3,19E-39 -0,1214539 1,59E-22 0,17046541 4,71E-43 -0,1854501 9,16E-51 0,13165274 3,25E-26 0,12507476 8,44E-24 -0,0990854 1,77E-15 -0,0903862 4,01E-13 -0,1541852 1,91E-35 -0,1772246 1,91E-46 -0,1865681 2,29E-51 -0,0258827 0,03811346 0,10436868 5,15E-17 -0,1582907 2,74E-37 -0,0778394 4,25E-10 -0,1087184 2,45E-18 -0,1493312 2,49E-33 -0,0194655 0,11890246 -0,0637726 3,64E-07 0,014704 0,23883695 0,14991399 1,40E-33 -0,1290439 3,05E-25 0,13738529 2,02E-28 0,11240874 1,67E-19 -0,1635813 9,72E-40 -0,2216094 3,03E-72 -0,1612042 1,26E-38 -0,0674442 6,35E-08 0,11721381 4,44E-21 Pareto’s Correlation p-value 0,24352551 2,64E-87 -0,0313998 0,01187497 0,16929654 1,76E-42 0,03533975 0,00463003 0,01222568 0,32740674 0,10614443 1,51E-17 0,07554268 1,36E-09 -0,1164197 8,17E-21 0,11684958 5,88E-21 -0,0729914 4,78E-09 -0,0713802 1,03E-08 0,04328502 0,00052262 0,04427245 0,00038802 0,09165888 1,87E-13 0,12256121 6,54E-23 0,13824015 9,28E-29 0,02089427 0,09415467 -0,0615421 8,04E-07 0,09582962 1,43E-14 0,06701184 7,71E-08 0,05555651 8,44E-06 0,09655145 9,03E-15 0,03211545 0,01007602 0,01210397 0,33478269 0,00454284 0,71593498 -0,1083106 3,27E-18 0,07132119 1,06E-08 -0,0764029 8,84E-10 -0,098463 2,65E-15 0,10566652 2,10E-17 0,14889402 3,83E-33 0,10384871 7,36E-17 0,04010906 0,00130831 -0,0758892 1,15E-09 2 Figure A1. Tree plot resulting from the Partition analysis performed considering the slope of the normalized biomass-size spectra (NBSS) as the response variable, and all the topological network parameters of the food-webs as predictor factors (only the variables chosen by the analysis are shown). Overall rNBSS2 = 0.594. 3 Figure A2. Tree plot resulting from the Partition analysis performed considering Pareto’s shape parameter as the response variable, and all the topological network parameters of the food-webs as predictor factors (only the variables chosen by the analysis are shown). Overall rPareto2 = 0.519. 4 Some definitions and formulations related to the model The food-web models were built considering virtual allometrically derived species (VADS), i.e. aggregation of individuals with the same body size, independent of their taxonomy, and sharing the same set of predators and prey. A top VADS is a predator that has no predator. An intermediate VADS is a VADS that is both a predator and a prey. A basal VADS is a prey that has no prey. The numbers of basal, intermediate, top, and all VADS in a web are denoted by b, i, t, and S. Cannibalism (Can) is the act of one individual of a VADS consuming all or part of another individual of the same VADS as food. Size structured cannibalism, in which large individuals consume smaller conspecifics, is more common. Omnivores (Omn) is the fraction of VADS that consume two or more VADS and have food chains of different lengths (where a food chain is a linked path from a non-basal to a basal VADS). Herbivores (Herb) is the fraction of VADS that feed exclusively on basal VADS. Carnivores (Carn) is the fraction of VADS that feed exclusively on non-basal species. The number and proportion of top (t/S), intermediate (i/S), basal (b/S), herbivores (Herb/S), carnivores (Carn/S), omnivores(Omn/S) and cannibals (Can/S) are calculated considering that for all the cases the total number of VADS is S. We calculated a trophic level measure called the mean ‘short-weighted trophic level’ (TL) (Williams & Martinez 2004). For a particular taxon, short-weighted 5 trophic level is the average of ‘prey-averaged trophic level’ (1 plus the mean trophic level of all the taxon’s trophic resources) and ‘shortest trophic level’ (1 plus the shortest chain length from the consumer taxon to a basal taxon). S TL j 1 lij i 1 TLi nj where nj is the number of prey species in the diet of species j, lij represents the links between the species i and j in a food web with S species. A trophic link (hereafter, link) is any reported feeding or trophic relation between two VADS in a web. For example, a link between a top and an intermediate VADS is denoted as t-i and a link between two intermediate VADS as i-i. Connectance (C) is the proportion of realized links within the food webs, calculated as the number of actual links (L) divided by the squared number of VADS (S), which gives the number of possible links within a network. CL S2 This is considered a standard measure of food-web trophic interaction richness, as well as the links per VADS (L/S), which equals the mean number of VADS’ predators plus prey, also referred to as link density. Other concepts related to the connectance are the Clustering coefficient (Cl), i.e., the average fraction of pairs of VADS one link away from a VADS that are also linked to each other, and the SD linkedness, i.e., the variation of the overall number of links. 6 The standard deviation of mean generality (GenSD) and vulnerability (VulSD), quantify the respective variabilities of VADS' normalized prey (Gi) and predator (Vi) counts (Williams & Martinez 2000). Gi 1 s a ji L j 1 S Vi 1 s aij L j 1 S Normalizing with L/S makes standard deviations comparable across different webs by forcing mean Gi and Vi to equal 1. Functional Response type: Several function response models have been proposed, all of these models have deficiencies that affect their utility (Gutierrez 1996). We employ here the Holling model that considers a (constant) population of N identical individuals which can engage in only two activities: searching for food and handling a recently found item (Gurney & Nisbet 1998). It is assumed that while an organism is handling a food item it cannot continue to search, so the length of time required to process each item sets an upper limit to the rate at which food can be consumed. Holling argued that as prey density increases, search becomes trivial, and handling (time the predator spends pursuing, subduing and consuming each prey item it finds, and then preparing itself for further search) takes up an increasing proportion of the predator’s time (Begon, Mortimer & Thompson 1996). Predator Interference (c) is the degree to which individuals within population i interfere with one another’s consumption activities, which reduces i’s per capita consumption if c>0 (Brose et al. 2006). If there are many predators, all concentrated within profitable patches, they remove prey rapidly thus reducing the profitability of those patches. Thus, in general, we can expect the apparent attacking efficiency to decrease as predator density increases (Begon et al. 1996) 7 Carrying capacity: carrying capacity of logistic growth: the population size at which competition is so great, and net reproductive rate so modified, that the population replace itself each generation (Begon et al. 1996). Biological rates: the biological rates of production, R, metabolism, X, and maximum consumption, Y, follow negative-quarter power–law relationships with the species’ body masses: RP = a y M P0.25 X C =a x M −C0. 25 Y C =a y M −C0 . 25 where ar, ax and ay are allometric constants and the subscripts C and P indicate consumer and producer parameters, respectively (Yodzis and Innes 1992). The time scale of the system was defined by normalizing all rates according to the growth rate of the basal population, and the maximum consumption rates were normalized by the metabolic rates: r i= 1 X a M xi C x C RP ar M P yi= 0.25 Y C ay = X C ax Inserting these three equations into eqs. 2 and 3 (of Main Document) yields a population dynamics model with allometrically scaled parameters. The constants used (yj = 4 for ectotherm vertebrates and yi = 8 for invertebrates; eij = 0.85 for carnivores and eij = 0.45 for herbivores; K = 1; ar = 1; ax = 0.314 for invertebrates 8 and ax = 0.88 for ectotherm vertebrates) are consistent with prior studies (Brose et al. 2006; Otto et al. 2007; Rall et al. 2007; Brose 2008). References Begon M, Mortimer M, Thompson D (1996) Population Ecology: A Unified Study of Animals and Plants. 3rd edn. Wiley-Blackwell, UK. Brose U (2008) Complex food webs prevent competitive exclusion among producer species. Proc R Soc Lond B Biol Sci 275:2507–2514. Brose U, Williams RJ, Martinez ND (2006) Allometric scaling enhances stability in complex food webs. Ecol Lett 9:1228-1236. Gurney WSC, Nisbet RM (1998) Ecological Dynamics. Oxford University Press, USA. Gutierrez AP (1996) Applied Population Ecology: A Supply/Demand Approach. John Wiley & Sons, New York, USA. Otto SB, Rall BC, Brose U (2007) Allometric degree distributions facilitate food-web stability. Nature 450:1226–1229. Rall BC, Guill C, Brose U (2007) Food-web connectance and predator interference dampen the paradox of enrichment. Oikos 117(2):202–213. Williams RJ, Martinez ND (2000) Simple rules yield complex food webs. Nature 404:180–183. Williams RJ, Martinez ND (2004) Limits to trophic levels and omnivory in complex food webs: theory and data. Am Nat 163:458-468. Yodzis P, Innes S (1992) Body size and consumer-resource dynamics. Am Nat 139:1151–1175. 9