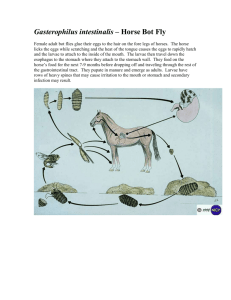
Materials and Methods
advertisement

Drosophila growth-blocking peptide-like factor mediates acute immune reactions during infectious and non-infectious stress Seiji Tsuzuki1, Masanori Ochiai2, Hitoshi Matsumoto1, Shoichiro Kurata3, Atsushi Ohnishi4 and Yoichi Hayakawa1* 1 Department of Applied Biological Sciences, Saga University, Honjo 1, Saga 840-8502, Japan, 2Institute of Low Temperature Science, Hokkaido University, Sapporo 060-0819, Japan, 3Graduate School of Pharmaceutical Sciences, Tohoku University, Sendai 980-8578, Japan, 4The Institute of Physical and Chemical Research, Wako351-0198, Japan Supplementary Information Materials and Methods Chemicals - Peptides such as Bombyx GBP (paralytic peptide), Lucilia GBP, and Drosophila GBPs were synthesized by the solid phase method using standard Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry. Synthesized peptides were suspended at a concentration of 1 mg/ml in 10 mM Tris-HCl, pH8, and stored at 4oC for 15-20 h. After checking disulfide bond formation by reversed-phase HPLC with a C18 column (250 x 4.6 mm, RP-18, Kanto Chemical Co.), the peptide samples were purified by HPLC with a preparatory C18 column (250 x 10 mm, RP-18, Kanto Chemical Co.) as described previously1. Peptides were solubilized in NaCl/Pi buffer (8 mM Na2HPO4, 1.5 mM KH2PO4, 137 mM NaCl, and 2.7 mM KCl, pH 7.2) when used in bioassay. Preparation of Hemolymph - The bluebottle fly hemolymph (approximately 0.7 ml) was collected in an ice cold microtube containing 0.7 ml of 50% acetone with 5 mM (p-amidinophenyl) methanesulfonyl fluoride hydrochloride (Wako, Japan) by cutting the anterior tip of the larvae. After centrifugation at 4oC for 10 min at 20,000 g, the supernatant was concentrated by lyophilization and the concentrated supernatant was used for further purification. Hemocyte Collection and in Vitro Bioassay - Hemolymph was collected from full-grown last instar larvae into anticoagulant solution (98 mM NaOH, 186 mM NaCl, 17mM Na2EDTA and 41 mM citric acid, pH 4.5) as described previously2. After a 1 30-min incubation on ice, hemocytes were collected by centrifugation at 4oC for 1 min at 300 g. The collected hemocytes were washed twice in Ex-cell 405 medium by sedimentation and resuspension. The ability of each fraction to induce hemocyte aggregation was assayed using 96-well culture plates (BM Equipment Co.). Plates were prepared by first adding 5 µl of the indicated sample solution to each well. Wells were then filled with 45 µl of the medium containing 1 x 104 hemocytes. Hemocytes were monitored by phase-contrast microscopy using an Olympus inverted microscope IX70. The activity was quantified by end-point dilution using the hemocyte aggregation assay. Bioassays with each sample were always paired with assays of bovine serum albumin (BSA) to control any variation in aggregation response that might exist between hemocyte samples2. Measurement of antibacterial activity of Bombyx mori larval hemolymph - Antibacterial activity was assayed by agar well diffusion method using Serratia marcescens. Wells of 5 mm diameter were punched in LB agar plates seeded with E. cloacae and the plates were then incubated at 37°C for 24 h. The diameter of a zone of clearing in the turbid agar around each well containing test hemolymph samples was measured to obtain a semi quantitative determination of the concentration of the antibacterial compound. Peptide Purification - The concentrated hemolymph sample was filtered through a membrane with an exclusion limit, 5,000 nominal molecular weight (Ultrafree-MC, Millipore Co.), and the filtrate was chromatographed by a C18 HPLC column (150 x 4.6 mm, UG120, Shiseido Co.) with a linear gradient of 0 - 50% CH3CN in 0.05% trifluoroacetic acid at a flow rate of 0.5 ml/min. The desired fraction was collected and further resolved by a C4 HPLC column (250 x 4.6 mm, YMC Co.) with a gradient of 10 40% CH3CN in 0.05% trifluoroacetic acid at a flow rate of 0.5 ml/min. Further purification was performed by a cyanopropyl-derived silica HPLC column (250 x 4.6 mm, UG120, Shiseido Co.) with a gradient of 10 - 40% CH3CN in 0.05% trifluoroacetic acid at a flow rate of 0.5 ml/min. The active fraction was resolved by a C 18 column (250 x 4.6 mm, RP-18, Kanto Chemical Co.) with a gradient of 15 - 40% CH3CN in 0.05% trifluoroacetic acid at a flow rate of 0.5 ml/min. Further, the active fraction was rechromatographed by the two tandem connected C18 columns (C18 - C18 column) (500 x 4.6 mm, RP-18(H), Kanto Chemical Co.) with a gradient of 15 - 30% CH3CN in 0.05% trifluoroacetic acid at a flow rate of 0.3 ml/min. The active fraction was finally purified to 2 homogeneity by the same column with a linear gradient of 18 - 28% CH3CN in 0.05% trifluoroacetic acid at a flow rate of 0.3 ml/min. The purified peptide was carboxymethylated and purified for sequencing. The carboxymethylated peptide was analyzed by automated Edman degradation with a protein sequencer (PPSQ 21, Shimadzu). The sequence was verified by analyzing approximately 100 pmol of the purified peptide twice. The sequenced peptide was synthesized by the solid phase method and their biological activities such as hemocyte aggregation and cell growth were tested. cDNA Synthesis and Cloning - Total RNA was extracted from the bluebottle fly larval fat body using TRIzol (Gibco-BRL) according to the manufacturer’s protocol. For cDNA syntheses, polyadenylated mRNAs were purified from total RNA using the QuickPrep mRNA purification kit (GE Healthcare). A cDNA fragment encoding the bluebottle fly cytokine was amplified by PCR using the following degenerate primers designed from the determined peptide sequence: GCNCCNTCNAAYTGYC and RCANCKNCCYTTRAA. Afterwards, by repeating 5’- and 3’-RACE methods, a cDNA fragment containing an entire ORF of the bluebottle fly cytokine was sequenced (AB243069). RT-PCR Analysis - To determine whether the Drosophila melanogaster genes are expressed in each species of larvae, RT-PCR was conducted essentially according to the procedure described previously3. First-strand cDNA was synthesized with oligo(dT)12-18 primer using ReverTra Ace RT-PCR kit (Toyobo) according to the manufacturer’s protocol. PCR amplification was performed with the following primers using a profile as follows: 35 cycles of 30 sec at 95oC, 1min at 55oC and 1.5 min at 72oC. For CG11395: TTTGCAGCCTCGACCCGG and GGCTTCCTTCCTGCAACG, for CG12517: ATGAGTAACCTGGGATCAATAC and ATGAACACAGCGACCACAATTG and ACTTGTGTCCAGCAGAATTTC, for CG14069: GAATCTCGAGTAGCATTTCACC, for CG15917: ATGTTGATACGTATTAATCCATTG and TTACGCCGGCTTTCTGCATC, for CG17244: CCAGTCCCGTTCAGCCCA and CCCGACAGCGACCACGAT. PCR analysis of CG15917 gene expression in the transgenic fly was performed with the above primers using a modified profile as follows: 25 cycles of 30 sec at 95oC, 1 min at 55oC and 1 min at 72oC. 3 Real-time quantitative PCR analysis of AMP gene expression in B. mori and D. melanogaster larvae was carried out by using the Light-Cycler 1.3 instrument and software (Roche Applied Science) as described previously3. PCR specificity was confirmed by the molecular masses of the PCR products and melting curve analysis at each data point. The copy numbers of RNA coding the genes of interest were standardized against that of the RNA coding rp49 in each sample. Specific primer pairs of Drosophila AMPs are described previously4 and those of Bombyx AMPs are as follows. Alpha-tubulin: ACTAACTTGGTGCCTTACCC and ACTAACTTGGTGCCTTACCC, Cecropin-A: CCGAGCACTATAGAATTTCGG and TCGCTTGCCCTATGACGGCTA, Moricin-1: GCAAAAACAGTAAACCGCGCAG and AAACATCGTTGGCTGTACTGG, Attacin: CAGACAAGTAATACGACACAGG and ATGGCGCTGAGCACGTTCTTGT, Gloverin-1: ACGCAGAAGTTTACGGACCTTC and TGCCGCGGTCGTCATTAAAGAT. Drosophila eggs were collected for 10-12 h at 25oC from test female flies and, three or four days after collecting eggs, 5 third instar larvae were randomly selected and used for measurement of AMP expression levels in each experiments. AMP expression levels in test larvae were measured within 3 h after GBP overexpression by heat treatment. We further confirmed no significant difference in Mtk and Dpt expression levels between day 3 and day 4 larvae. Therefore, under these experimental conditions, the GBP-induced delay in larval development could not affect AMP expression levels. References 1. Ohnishi, A., Oda, Y. & Hayakawa, Y. Characterization of receptors of insect cytokine, growth-blocking peptide, in human keratinocyte and insect Sf9 cells. J. Biol. Chem. 41, 37974-37979 (2001). 2. Aizawa, T. et al. Structure and activity of the insect cytokine growth-blocking peptide. J. Biol. Chem. 276, 31813-31818(2001). 3. Ninomiya, Y. et al. Insect cytokine growth-blocking peptide signaling cascades regulate two separate groups of target genes. FEBS J. 275, 894-902(2008). 4. Takehana, A. et al. Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial 4 defense and the prophenoloxidase cascade in Drosophila larvae. Proc. Natl Acad. Sci. USA 99, 13705-13710 (2002). 5. Levashina, E.A., Ohresser, S., Lemaitre, B. & Imler, J.L. Two distinct pathways can control expression of the gene encoding the Drosophila antimicrobial peptide metchnikowin. J. Mol. Biol. 278, 515-527 (1998). 5 Fig. 1 - Purification of Lucilia GBP from the Lucilia larval hemolymph Six analytical HPLC runs using reverse phase columns monitored at 220 nm in combination with hemocyte aggregation assays resulted in isolation of an active peptide. A, C18 active fraction is indicated by horizontal bar. B, C4 active fraction is indicated by horizontal bar. C, Cyanopropyl active fraction is indicated by horizontal bar. D, C18 active fraction is indicated by horizontal bar. E, C18-C18 active fraction is indicated by horizontal bar. F, C18-C18 active fractions is indicated by horizontal bar. 6 Fig. 2 - A 554 bp cDNA encodes Lucilia GBP homolog Nucleotide sequence of Lucilia pre-pro-GBP is described with the deduced amino acid sequence for the predicted protein shown below the corresponding codons. The underlined and red-colored sequences indicate the putative signal peptide sequence and active peptide sequence, respectively. 7 Fig. 3 - Sequence alignment of the GBP homolog genes Amino acid sequence alignment of genes encoding GBP and the homologous peptides identified in Lepidoptera (Pseudaletia separata, Mamestra brassicae, and Spodoptera litura) and Diptera (Lucilia cuprina and Drosophila melanogaster). Red-colored sequences at C-terminal ends indicate active GBPs (or putative active GBPs after processing by a serine proteinase). Conserved cysteine residues are emphasized by green bands. The number in parenthesis indicates the total number of amino acid residues found in the open reading frame of each gene. 8 Fig. 4 - Timing of pupariation of third instar larvae of transgenic Drosophila larvae Same-staged larvae of control lines (hs-Gal4 and UAS-GBP) and GBP overexpression line (hs-Gal4/UAS-GBP) were heated at 33oC for 30 min. 9 Fig. 5 - Expression of the GFP reporter gene in Mtk-GFP transgenic larvae (a) Mtk-GFP transgenic larvae5 without any treatment. (b) Mtk-GFP transgenic larvae treated at the heat treatment, 35oC for 30 min. (c) Expression of the Mtk-GFP reporter in transgenic larvae (UAS-proGBP/+; hs-GAL4/ Mtk-GFP) by the forced expression of proGBP under the direction of an hs-Gal4 driver after the heat treatment. (d) Expression of the reporter in the larvae stabbed with a needle previously dipped in 100 µM dGBP. (e) Expression of the reporter in the larvae stabbed with a sterile needle. (f) Expression of the reporter in the larvae stabbed with a needle coated with Gram-negative bacteria (Serratia marcescens). GFP expression was recorded at 6 hr after the heat treatment for proGBP overexpression or stabbing with a needle. 10 Fig. 6 - Effects of GBP overexpression driven by an actin-Gal4 driver on Mtk, Dpt, and Drs expression in Drosophila adults Metchnikowin (Mtk), Diptericin (Dpt), and Drosomycin (Drs) expression levels were measured in Drosophila adults with overexpression of GBP under the direction of an actin-Gal4 driver. Data are given as means + S.D. for three separate measurements. Asterisks indicate significant differences from control UAS-GBP line (t-test; *P<0.05, **P<0.01). 11 Fig. 7 - Reduction of Drosophila GBP expression in GBP RNAi larvae after heat treatment (a) RT-PCR was performed by using total RNAs prepared from whole bodies of GBP RNAi larvae at indicated times after heat treatment. (b) Hemolymph peptide fraction was prepared from GBP RNAi larvae at indicated times after heat treatment, and used for Western blot analysis. Peptide fraction was prepared in the same way as hemolymph peptides of the bluebottle fly larvae (see Materials and Methods). Note that 36 h after heat treatment, no signal could be detected by anti-GBP antibody. (c) Fat bodies isolated from control and GBP RNAi larvae 36 h after heat treatment. (d) Weight ratio of fat body to the whole body (white bar) and protein concentration in fat body (gray bar) of each test animal (control and GBP RNAi lines). Wet weight of fat body was measured soon after absorption of excessive water. Data are given as means + S.D. for four separate measurements. Note that these results including (c) and (d) indicate the normality of fat body in GBP RNAi larvae. 12 Fig. 8 - Bacteria growth in GBP RNAi larvae after injection GBP RNAi and control larvae were challenged with Gram-negative bacteria Serratia marcescens and 1 µl of hemolymph was collected from each larvae 6 h after the bacterial injection. The collected hemolymph was diluted serially and plated onto a LB agarose plate. S. marcescens CFUs were determined by testing four groups of five larvae for each time point. Data are given as means + S.D. for four separate measurements. *, P<0.05 versus control by t-test. 13 Fig. 9 - Survival rate of GBP RNAi larvae after aseptic injury Control (●) and GBP RNAi larvae (○) were stabbed with a sterile needle (diameter: approximately 0.23 mm) and cultivated in a sterile tube containing autoclaved food. The survival rates were measured at indicated times. Data are given as means + S.D. for three separate measurements. Significant difference between control and test larval slopes were determined using ANOVA with the general linear model procedures of Minitab (release 14, Minitab Inc., USA). 14 Fig. 10 - GBP overexpression in bsk RNAi larvae Bsk and GBP expression levels were measured by using total RNAs prepared from control and test Drosophila larvae. Note that bsk expression was apparently repressed but GBP was overexpressed in UAS-GBP/+;actin-Gal4/UAS-dsbsk larvae. 15 Fig. 11 - GBP-dependent JNK phosphorylation in Drosophila larvae and S2 culture cells (a) Western blot analysis of the phosphorylation of fat body JNK (p-JNK) in Drosophila larvae with GBP overexpression. Note that phosphorylation levels of fat body JNK was significantly increased 1 h and 2 h after heat treatment at 35oC for 30 min. BH: before heat treatment; 0 h: immediately after heat treatment. (b) Western blot analysis of the phosphorylation of JNK in Drosophila S2 cells. Note that GBP-induced phosphorylation of JNK was clearly observed in S2 cells 10 min after adding 100 nM synthetic GBP into the culture medium. 16