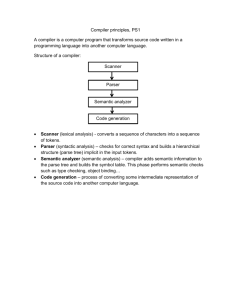
CHARMM Element doc/genborn $Revision: 1.4 $ File: GBORN
advertisement

CHARMM Element doc/genborn.doc $Revision: 1.4 $
File: GBORN, Node: Top, Up: (chmdoc/commands.doc), Next: Syntax
Generalized Born Solvation Energy and Forces Module
The GBORN module permits the calculation of the Generalized Born
solvation energy and forces of this energy following a formulation
similar to that of Still and co-workers and as described in the
manuscript from B. Dominy and C.L. Brooks, III (see below). This
module implements the following equation for the polarization energy,
Gpol:
G
=
-C
q q
N
N
i j
(1-1/eps){1/2 sum sum ------------------------------------
}
pol
el
i=1 j=1 [r^2 + alpha *alpha exp(-D )]^(0.5)
ij
i
j
ij
The gradient of the function is also computed so forces due to solvent
polarization can be utilized in energy minimization and dynamics. In
its current implementation, the calculation of the alpha(i) variables
and the sums over particles indicated in the sums above are done
without cutoffs, therefore for large systems these can be costly
calculations (though still less so than for explicit solvent).
Questions and comments regarding implementation of these equations or
there parameterization for the CHARMM forcefields (param19/toph19,
param22 for proteins and nucleic acids) should be directed to Charles
L. Brooks, III at brooks@scripps.edu. Use of the GB term for MMFF and
CFF has recently been implemented and the parameters are given below
under examples.
The appropriate citation for this work is:
B. Dominy and C. L. Brooks, III. Development of a Generailzed Born
Model Parameterization for Proteins and Nucleic Acids.
J. Phys. Chem., 103, 3765-3773(1999).
An alternative method for calculating atomic Generalized Born radii
(alpha values) is available. This method extends the original
formalism by Still and coworkers and has atom type specific
parameters. The resulting solvation free energies are generally
more accurate with respect to reference Poisson-Boltzmann energies.
So far parameter sets are only available for CHARMM19 and CHARMM22
protein models.
* Menu:
* Syntax::
Syntax of the GBORN Commands
* Function::
* Examples::
Purpose of each of the commands
Usage examples of the GBORN module
File: GBORN, Node: Syntax, Up: Top, Previous: Top, Next: Function
Syntax of Generalized Born Solvation commands
[SYNTAX: GBORn commands]
GBORn { ( P1 <real> P2 <real> P3 <real> P4 <real> P5 <real> [LAMBda
<real>] |
GBTYpe 10 PARUnit <int> )
[LAMBda <real>] [EPSILON <real>] [CUTAlpha <real>] [WEIGht]
[ANALysis] }
{ CLEAr
}
File: GBORN, Node: Function, Up: Top, Previous: Syntax, Next: Examples
Parameters of the Generalized Born Model in the original Still
model
P1-P6
The parameters P1, P2, ..., P5 specify the particular
parameters
controlling the calculation of the effective Born radius for
a particular configuration of the biomolecule as determined
from
the sum over all atoms:
Alpha(i) = [-CCELEC/(2*Lambda*R(i)) + P1*CCELEC/(2*R(i)^2
+ Sum {P2*V(j)/rij^4} + Sum {P3*V(j)/rij^4}
bonds
angles
+ Sum {P4*V(j)*Cij/rij^4}]^(-1)*(-CCELEC)/2
non-bonded
with Cij = 1 when (rij^2)/(R(i)+R(j))^2 > 1/P5
and Cij = 1/4(1-cos[P5*PI*(rij^2)/(R(i)+R(j))^2])^2 otherwise.
Note:
R(i), V(i) correspond to the vdW radius and volume respectively,
CCELEC is the conversion from e^2/A to kcal/mol, rij is the
separation
between atom i and atom j.
Lambda
term
This is the scaling parameter for the vdW radius in the first
of the expression above.
Note:
The parameters P1-P5 and Lambda correspond to parameters for a
particular CHARMM parameter/topology set.
***The parameters P1-P5 and Lambda are required input***
Parameters of the Generalized Born Model in the extended approach
GBTYpe 10
selects the extended approach for calculating Generalized
Born radii
PARUnit
read
is used to specify the unit from which the parameter set is
If this option is not given parameters are read from the
current
input stream.
The parameter set is expected to contain a single line
for for each atom type that occurs in the modeled structure
with the following values:
Atomtype Pr0 Pb Pa Pn Pmf Pef Pgd Pgn Pdc
They are used as follows in the calculation of GB radii:
G(i) = Pr0 * 1/R(i) + Pb * Sum V(j)/rij^4 +
bonds
Pa * Sum V(j)/rij^4 * CCF +
angles
Pnb * Sum V(j)/rij^4 * CCF
non-bond
F(i) = G(i) * ( 1 + Pmf * Sum
V(j)/rij^3 )
non-bond
Alpha(i) = 1 / ( Pgd + F(i) + Pef * F(i)*F(i) ) + Pgn
Pdc is used to modify
Dij in the GB approximation to:
Dij = rij^2 / ( (Pdc(i) + Pdc(j)) * Alpha(i) * Alpha(j) )
The parameter set needs to be terminated with a line
containing only 'END'.
Common Parameters for both models
EPSILON
medium.
This is the value of the dielectric constant for the solvent
The default value is 80.
CUTAlpha
This is a maximum value for the effective Alpha for any atom
during the calculation for a particular conformation of the
biomolecule. It is necessary because in some instances the
expression above for Alpha(i) can take on negative values
of numerical problems with the expression for very buried
atoms
in large globular biopolymers.
10^6.
WEIGht
The default for this value is
This is a flag to specify that you want the vdW radii for the
atoms to be taken from the wmain array instead of the
parameter
files (from Rmin values). The default is to use the parameter
values. These values are used for the R(i) and V(i) noted
above.
ANALysis
This flag turns on an analysis key that puts the atomic
contributions
to the Generalized Born solvation energy into an atom array
(GBATom)
for use through the scalar commands.
CLEAr
Clear all arrays and logical flags used in Generalized Born
calculation.
FEP calculations with the original Still model
GBTYpe
GBTYpe permits GB energy calculation with block module.
Environmental atoms should be assigned to block 1. The
variable
parts are assigned to blocks n (n > 1).
GB energy in the intermediate state can be expressed in two
ways.
Therefore, we can choose type 1 or 2. In common, Type 1 is
computationally inexpensive and extensible. In particular,
the computational time increases rapidly with GBTYpe=2 as
the number of blocks increases. When GBYTyp is used, block
command
also should be used.
Note:
GBTYpe allows the use of GB energy with FEP calculations
(BLOCK module), lambda-dynamics method, hybrid-MC/MD, and
replica.
Typical input examples can be found in testcase of Version 28.
The definintions of Type 1 and 2 are shown next.
Type 1
/
q q
q q
q q
\
| env env
ligk ligk
i j
|
i j
L
2
env ligk
i j
G
= -C (1-1/eps)1/2| sum sum------ + sum lambda (2 sum sum ------ +
sum sum -------|
pol
el
| i
j F
k=1
k
i
j
F
i
j
F
|
\
ij
ij
ij /
F
ij
= [r^2 + alpha *alpha exp(-D )]^(0.5)
ij
i
j
ij
(1) When ith atom belongs to environmental atoms
Alpha
= [-CCELEC/(2*Lambda*R(i)) + P1*CCELEC/(2*R(i)^2
i
env
env
+ Sum {P2*V(j)/rij^4} + Sum {P3*V(j)/rij^4}
bonds
angles
env
+ Sum {P4*V(j)*Cij/rij^4}]^(-1)*(-CCELEC)/2
non-bonded
L
2
+ sum lambda
k=1
k
/ ligk
ligk
|+ Sum {P2*V(j)/rij^4} + Sum {P3*V(j)/rij^4}
\ bonds
angles
ligk
\
+ Sum {P4*V(j)*Cij/rij^4}]^(-1)*(-CCELEC)/2 | ]
non-bonded
/
(2) When ith atom belongs to ligand k
Alpha
= [-CCELEC/(2*Lambda*R(i)) + P1*CCELEC/(2*R(i)^2
i
env
env
+ Sum {P2*V(j)/rij^4} + Sum {P3*V(j)/rij^4}
bonds
angles
env
+ Sum {P4*V(j)*Cij/rij^4}]^(-1)*(-CCELEC)/2
non-bonded
ligk
ligk
+ Sum {P2*V(j)/rij^4} + Sum {P3*V(j)/rij^4}
bonds
angles
ligk
+ Sum {P4*V(j)*Cij/rij^4}]^(-1)*(-CCELEC)/2
non-bonded
]
with Cij = 1 when (rij^2)/(R(i)+R(j))^2 > 1/P5
and Cij = 1/4(1-cos[P5*PI*(rij^2)/(R(i)+R(j))^2])^2 otherwise.
Type 2
/
q q
q q
q q
\
L
2| env env i j
env ligk i j
ligk
ligk i j |
G
= -C (1-1/eps)1/2 sum lambda| sum sum----- + sum sum ------ + sum
sum -------|
pol
el
k=1
k| i
j
F(k)
i
j
F(k)
i
j
F(k) |
\
ij
ij
ij /
F(k) = [r^2 + alpha(k) *alpha(k) *exp(-D )]^(0.5)
ij
ij
i
j
ij
(Each environmental atom has the L Born radius)
Alpha(k) =
i
[-CCELEC/(2*Lambda*R(i)) + P1*CCELEC/(2*R(i)^2
env
env
+ Sum {P2*V(j)/rij^4} + Sum {P3*V(j)/rij^4}
bonds
angles
env
+ Sum {P4*V(j)*Cij/rij^4}]^(-1)*(-CCELEC)/2
non-bonded
ligk
ligk
+ Sum {P2*V(j)/rij^4} + Sum {P3*V(j)/rij^4}
bonds
angles
ligk
+ Sum {P4*V(j)*Cij/rij^4}]^(-1)*(-CCELEC)/2
non-bonded
]
with Cij = 1 when (rij^2)/(R(i)+R(j))^2 > 1/P5
and Cij = 1/4(1-cos[P5*PI*(rij^2)/(R(i)+R(j))^2])^2 otherwise.
File: GBORN, Node: Examples, Up: Top, Previous: Function, Next: Top
Examples
The examples below illustrate some of the uses of the generalized Born
model. See c27test/genborn19.inp, c27test/genborn22.inp
See c28test/gbmf19.inp for examples on how to use the extended approach
for
calculating atomic Generalized Born radii.
Example (1)
----------Calculate the generalized Born solvation energy using atomic radii from
the
wmain rray (example illustrates the useage but simply uses the same radii
as
would be employed w/o the "Weight" option). Using a switching function
for
the solvation and electrostatice between 14 and 18 A.
! Test use of radii from wmain array
scalar wmain = radii
! Now turn on the Generalized Born energy term using the param19
parameters
GBorn P1 0.4153 P2 0.2388 P3 1.7488 P4 10.4991 P5 1.1 Lambda 0.7591 Epsilon 80.0 Weight
! Now calculate energy w/ GB
energy cutnb 20 ctofnb 16 ctonnb 14
GBorn Clear
Example(2)
---------Calculate the generaized Born solvation energy and use the ANALysis key
to
access atomic solvation energies.
GBorn P1 0.4153 P2 0.2388 P3 1.7488 P4 10.4991 P5 1.1 Lambda 0.7591 Epsilon 80.0
mini sd nstep 1000
!!!!CHECK SCALAR RECALL of GB variables
! What are the current Generalized Born Alpha, SigX, SigY, SigZ and T_GB
! and atomiuc solvation contribution (GBATom) values?
skipe all excl GbEnr
energy cutnb @cutnb
scalar GBAlpha show
scalar SigX show
scalar SigY show
scalar SigZ show
Scalar T_GB show
Scalar GBAtom show ! One can now use the individual contributions for
whatever.
GBorn Clear
Example(3)
---------Do a minimization (could be dynamics too, forces are computed exactly)
! Finally minimize for 1000 steps using SD w/ all energy terms.
skipe none
GBorn P1 0.4153 P2 0.2388 P3 1.7488 P4 10.4991 P5 1.1 Lambda 0.7591 Epsilon 80.0
mini sd nstep 1000 cutnb 20 ctofnb 18 ctonnb 14 switch
***Note: We find that the generailzed Born energy together with
electrostatics
converges quickly as a function of cutoff
Example (4)
----------Use of GB term with MMFF and CFF forcefields in CHARMM.
1.
Make sure CHARMM was compiled with CFF and/or MMFF keywords.
2. Commands are the same as above and may be used as with the CHARMM
forcefields.
3. Parameters for these systems are given below, taken from testcases
in c27test/GB_*.inp
-------------------------CFF95---------------------------------------***GENERAL
! Now turn on the Generalized Born energy term
! using the CFF95 general parameters
GBorn P1 0.4475 P2 0.4209 P3 0.0120 P4 8.4186 P5 0.9 Lambda 0.7660
Epsilon 80.0
***Single AA
! Now turn on the optimized generalized Born energy term
! for MMFF - lambda optimized for single AA
GBorn P1 0.4475 P2 0.4209 P3 0.0120 P4 8.4186 P5 0.9 Lambda 0.7703
Epsilon 80.0
***dipeptides
! Now turn on the optimized generalized Born energy term
! for CFF95 - dipeptide optimized.
GBorn P1 0.4475 P2 0.4209 P3 0.0120 P4 8.4186 P5 0.9 Lambda 0.7686
Epsilon 80.0
***Proteins
! Now turn on the optimized generalized Born energy term
! for CFF95 - optimized for proteins.
GBorn P1 0.4475 P2 0.4209 P3 0.0120 P4 8.4186 P5 0.9 Lambda 0.6957
Epsilon 80.0
***NA bases
! Now turn on the optimized generalized Born energy term
! for CFF95 - lambda optimized for NA base
GBorn P1 0.4475 P2 0.4209 P3 0.0120 P4 8.4186 P5 0.9 Lambda 0.7682
Epsilon 80.0
***Di-NAs
! Now turn on the optimized generalized Born energy term
! for CFF95 - lambda optimized for dinucleotides
GBorn P1 0.4475 P2 0.4209 P3 0.0120 P4 8.4186 P5 0.9 Lambda 0.7681
Epsilon 80.0
***NA strands
! Now turn on the optimized generalized Born energy term
! for CFF95 - lambda optimized for NA strands
GBorn P1 0.4475 P2 0.4209 P3 0.0120 P4 8.4186 P5 0.9 Lambda 0.7461
Epsilon 80.0
-------------------------MMFF---------------------------------------***GENERAL
! Now turn on the optimized generalized Born energy term
! for MMFF - generic w/o molecule-based lambda optimized.
GBorn P1 0.2163 P2 0.2564 P3 0.0144 P4 7.0038 P5 1.0 Lambda 0.91 Epsilon
80.0
***Small organics
! Now turn on the optimized generalized Born energy term
! for MMFF small molecules.
GBorn P1 0.2163 P2 0.2564 P3 0.0144 P4 7.0038 P5 1.0 Lambda 0.91 Epsilon
80.0
***Single AA
! Now turn on the optimized generalized Born energy term
! for MMFF - lambda optimized for single AA
GBorn P1 0.2163 P2 0.2564 P3 0.0144 P4 7.0038 P5 1.0 Lambda 0.8874
Epsilon 80.0
***dipeptides
! Now turn on the optimized generalized Born energy term
! for MMFF - lambda optimized for diaas
GBorn P1 0.2163 P2 0.2564 P3 0.0144 P4 7.0038 P5 1.0 Lambda 0.8649
Epsilon 80.0
***Proteins
! Now turn on the optimized generalized Born energy term
! for MMFF - lambda optimized for proteins
GBorn P1 0.2163 P2 0.2564 P3 0.0144 P4 7.0038 P5 1.0 Lambda 0.8417
Epsilon 80.0
***NA bases
! Now turn on the optimized generalized Born energy term
! for MMFF - lambda optimized for NA base
GBorn P1 0.2163 P2 0.2564 P3 0.0144 P4 7.0038 P5 1.0 Lambda 0.8787
Epsilon 80.0
***Di-NAs
! Now turn on the optimized generalized Born energy term
! for MMFF - lambda optimized for dinucleotides
GBorn P1 0.2163 P2 0.2564 P3 0.0144 P4 7.0038 P5 1.0 Lambda 0.8768
Epsilon 80.0
***NA strands
! Now turn on the optimized generalized Born energy term
! for MMFF - lambda optimized for NA strands
GBorn P1 0.2163 P2 0.2564 P3 0.0144 P4 7.0038 P5 1.0 Lambda 0.8281
Epsilon 80.0