gothenburg_protocol
advertisement

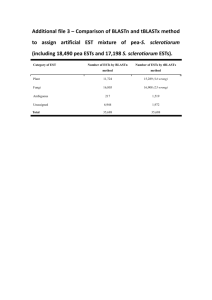
Summary of issues discussed during the second meeting of the “Sleeping Beauty” project. (8-9/5/2006) Present: P1 [Esther Lubzens, Ora Hadas, Nadav Denekamp (post-doc)]; P2 [Joan Cerdá, Angèle Tingaud-Sequeua (post-doc)]; P3 [Stefan Hohmann, Avi Nahmani (PhD student), Ivan Pirkov (PhD student), Cecilia Geijer (PhD student)]: P4 [Melody Clark, Jelena Pilipović (PhD student)], P5 [Richard Reinhardt], P6 [Kristian Fog Nielsen]. Full and updated contact list is attached to this protocol Notes were taken by Nadav Denekamp 1 Agenda Sleeping Beauty – Biennial Meeting, Novotel, Göteborg May 7-9, 2006 Organized by Dr. Stefan Hohmann, Cecilia Geijer Department of Cell and Molecular Biology/Microbiology Göteborg University Sweden cecilia.geijer@gmm.gu.se May 7: Meet on Sunday afternoon (15:00 Novotel Lobby) and go to Stefan's place for a social event and informal discussions. Transfer will be arranged. Return from Stefan’s place at 2000. In case of direct transfer from airport: Address: Norra häcksjöbäcksvägen 46, 44332 Lerum Tel: +46 (0)302 17689, +46 (0)733547297 1 May 8 – May 9 lunch time: presentations and discussions at a location to be specified in Göteborg. Presentations and Discussions 1. May 8: Presentations by partners and the co-ordinator. Partners will present the progress, obstacles and plans for the following six months for each part they participate in each one of the workpackages. Partner 6 (DTU) will present their institute and their role in the project. 2. May 9: Each leader of a work package will summarise the progress in their workpackage and highlight obstacles and plans for the forthcoming 6 months. Any deviation in the workplan or the timetable will be discussed. Discussions will be held on the flow of samples and their analyses by Partner 5 (MPI-MG) and Partner 6 (DTU). Discussions will be held on topics raised by partners. Discussions will be initiated on the Dormancy workshop in 2008. 2 1.1 Agenda for Monday May 8, 2006 0900 - 0915 Welcome Stefan Hohmann (P3) 0915 – 0945 Cynanobacteria akinetes Ora Hadas, Assaf Sukenik (P1-KLL) 0945 - 1015 Yeast spores Stefan Hohmann (P3) 1015 - 1045 Coffee Break 1045 - 1115 Rotifer resting eggs Esther Lubzens (P1NIO) 1115 - 1145 Arctic Springtails Roger Worland, Melody Clark (P4) 1145 - 1215 Killifish eggs Joan Cerdá (P2) 1215 -1245 Discussion Esther Lubzens 1245 - 1400 Lunch 1400 - 1530 Management and coordination of project Stefan data – Workpackage 5 and Melody Clark DTU and metabolomics Kristian Fog Nielsen 1530 - 1600 Hohmann (P6) 1600 - 1630 Molecular and Proteomic tools Richard Reinhardt (P5) 1630- 1700 Coffee break 1700 – 1800 Discussion on tools and General Discussion Richard Reinhardt, Kristian Fog Nielsen and Esther Lubzens 1900 Dinner 3 1.2 Agenda for Tuesday, May 9, 2006 0830 -0845 Inroduction into discussion on SB Esther Lubzens workplan and Workpackage 0 0845 - 0915 Workpackage 1 – Coordinators: Ora Hadas, Assaf Sukenik (P1Esther Lubzens & Ora Hadas KLL), Esther Lubzens (P1-NIO), Stefan Hohmann (P3), Richard Reinhardt (P5) 0915 - 0945 Workpackage 2 – Coordinator: Stefan Hohmann Ora Hadas, Assaf Sukenik (P1KLL), Esther Lubzens (P1-NIO), Stefan Hohmann (P3), Richard Reinhardt (P5) 0945 - 1015 Coffee Break 1015 – 1045 Workpackage 3 – Coordinator: Joan Cerdá Joan Cerdá (P2) Roger Worland (P4), Melody Clark (P4), Richard Reinhardt (P5) 1045 - 1115 Workpackage 4 – Coordinator: Joan Cerdá Esther Lubzens (P1-NIO), Joan Cerdá (P2), Stefan Hohmann (P3), Richard Reinhardt (P5) 1115 - 1145 Workpackage 5 – Conclusions from previous day Coordinators: Stefan Hohmann & All Partners Melody Clark 1145 -1215 Workpackage 6 – All partners Coordinator: Esther Lubzens 1215 -1230 Summing up 1230 Lunch Esther Lubzens 4 2 Presentations made by partners The presentations will be available in PDF format in the project web site. (http://www.gmm.gu.se/sleeping). The presentations included reports on achievements, deliverables and plans until M12 and for achieving milestones. 1. Cyanobacteria Dormancy Forms in an Aquatic environment [Ora Hadas] 2. Yeast spores 2.1. Sporulation in buddingyeast S. cerevisiae and the story of yeast aquaporin Aqy1 [Cecilia Geijer] 2.2. Mechanisms of Dormancy and Germination of The Baker’s Yeast S. cerevisiae Spore [Ivan Perkov] 3. Rotifer resting eggs [Nadav Denekamp and Esther Lubzens] 4. Arctic Sprintails [Melody Clark] 5. Killifish eggs [Angèle Tingaud-Sequeua] 3 Management and coordination of project data - WP5 [Stefan hohmann and Melody Clark] 3.1 Microarray experiments design and data analysis. Support for experiments design for those who do not have experience with microarray experiments (P1 and P2) will be given by P3 (i.e. Avi). Analysis of the microarray data will be done by each partner. A workshop on microarray analysis will be given by P3. 3.2 Analysis of the results Further analysis of the results (after data analysis) - discovery of common and divergent pathways for dormant stages, dormancy and desiccation resistance will be co-ordinated by P3 and P4 (Melody). It was suggested to build a platform to examine the effect of different conditions and/or stages within the life cycle on the expression of genes, however such platform maybe too complicated. 5 3.3 Common gene bank It was agreed that when the EST data (sequences and annotations) are available they will be linked to the project web site (privet area). The web site will contain a search and blast utilities within the ESTs of all partners. The issue of keeping the data confident was brought up. It was agreed that if one wishes to share data from the project with a collaborator he may share his/her own data but with permission of all partners. The main idea is to give the partners the privilege to use first knowledge that was generated from the project. When there are results of all the microarray experiments, there will be another discussion on the combination of the results. The discussion will take place in the end of the second year during the annual meeting. 4 DTU and metabolomics [Kristian F. Nielsen] The platforms offered by DTU are described at Kisrstian’s presentation, which will be available in the project website (PDF format). Partners should supply Kristian the following information: sample matrix (needed for sample preparation), amount available (mg, gram…), special metabolites and the number of samples. Partners will send samples to Kristian during the following months so that he will be able to develop methods for analysis. Please send an email to Kristian before sending samples. Kristian’s contact information is listed in the updated contact list attached to the protocol. 5 5.1 Molecular and proteomic tools [Richard] cDNA libraries and ESTs Until now P2(Joan) and P4(Melody) sent RNA samples to P5. P1-KLL (Ora) sent samples for genome sequencing but they need to be sent again. Samples fromP1-NIO (Esther) will be sent during the next 3 months. cDNA libraries will be constructed for different condition and/or stages. cDNA from all the libraries will be pooled into one normalized library. 6 Until the next meeting each partner should tell Richard how many additional ESTs (beyond the 15k covered by the project) are requested. The original list for Richard: 56K ESTs ~14 libraries Microarrays: 3,000 genes/array and 40 experimetns The current list: P1(Ora): Whole genome sequencing in exchange of 3 libraries and 14,000 ESTs. P1(Esther): About 6 libraries to be normalized into 1 and 16,000 ESTs P2(Joan): About 6 libraries to be normalized into 1 and 15,000 ESTs P4(Melody): 3(?) normalized libraries, no microarrays. The ESTs should be prepared until the end of 2006. 5.2 Microarrays Microarrays services of P5 are needed for P1 and P2. P4 (Melody) will make their own slides. Printing, hybridization and scanning of the first ten slides are covered within the project budget. Richard will supply the costs for additional hybridization experiments until the next meeting. The slides will be printed only once, thus, requests for additional slides should be made in advance. The available amount of cDNA may limit the number of slides. Every slide will contain 3,000 genes (including replicate spots). The slides may contain genes from one treatment or genes from two treatments (1,500 genes per treatment). This issue should be discussed in the next meeting. Hybridization and scanning of additional slides may be done in Berlin or by each partner. It was suggested to organize a workshop on hybridization experiments for those who whish to perform the experiments by themselves. 5.3 Proteome analysis Proteome analysis services of P5 are needed for P1, P2, P4. P3 (Stefan) will perform the proteome analysis for yeasts. 7 Proteome profiling: 2D gels and analysis of ~400 spots. 2D gels analysis needs optimization therefore partners are requested to send a picture of a 2D gel of their samples to Richard or to send samples just for optimization. The amount of protein needed for 2D gel is 50-100 g per lane. 6 6.1 Discussions on the workpackages. WP0 – management 6.1.1 Next meetings M12: meeting in Cambridge 16-18/10/2006. M18: Copenhagen DTU (May 2007). M24: Israel NIO+KLL. The place of the meeting will be decided according to a questionnaire that will be sent by email (October 2007). M30: The workshop on dormancy and the biennial meeting in Berlin (May 2008). M36: Barcelona (October, 2008). 6.1.2 Contract between the partners Esther will send a copy of a proposed contract between the partners. 6.1.3 Project website The website will be available soon. It was agreed not to put manuscript on the website but just the links to the publisher site. 6.2 WP1: Studies on mechanisms of establishments of dormant stages D4: Optimal conditions for the induction of akinetes in cyanobacteria, asexualy and sexually reproducing rotifers and their resting eggs (M9) – accomplished. D5: cDNA libraries , EST sequencing and database construction – in progress (M18). RNA samples from P1.NIO will be sent to P5 during the next 3 months. D6: Pattern of global gene expression during formation of akinetes, rotifer resting eggs by micorarray anaylysis (M27) – will start after D4 is accomplished. 8 D7: Quantitative analysis of expression of specific genes by QPCR (M35) – will start after D6 is accomplished. D8: Proteins and metabolites identification to be associated with formation of dormant stages within dormant forms (M30) – samples for metabolomics and proteomics calibration will be sent during the next months. D9: Genes, proteins and metabolites associated with the dormant stages of yeast spores (M36) – on work. 6.3 WP2: Studies on the mechanisms of arousal from dormant stage D10: Metabolic pathways associated with germination in akinetes and of vegetative reproducing cyanobacteria (M12) – in progress. D11: Pattern of global gene expression during exit from dormancy of akinetes , resting eggs and spores by microarray analysis (M35) – will start when D5 is finished. D12: Quantitative expriession levels of specific genes by QPCR (M35) – will start after D11 is finished. D13: Protein and metabolites identified to be associated with resume metabolic activities in akinetes and during embryonic development in resting eggs and asexual rotifer eggs (M30) - samples for metabolomics and proteomics calibration will be sent during the next months. D14: Genes, proteins and metabolites associated with yeast spores germination (M36) – on work. 6.4 WP3: Desiccation and revival D15: Identification of cooling rate that elicits trehalose synthesis in Arctic srpringtails (M12) – almost finished D16: Identification and characterization of killifish embryo stages resistant to air exposure (M12) – almost finished. 9 D17: Microarray analysis of global genes expression during desiccation resistance in sprintails and air exposure in killifish embryos – in progress, RNA samples were sent to P5. Results of ESTs of the killifish embryos should be available until October 2006. D18: ISH and quantitave mRNA expression levels of specific genes by Real-time PCR (M35) - QPCR for the genes HSP70, HIF and HIF2are in progress. D19: Protein and metabolic profiles associated with desiccated springtails and airexposed killifish embryos (M30) – samples for metabolomics and proteomics calibration will be sent during the next months. 6.5 WP4 - Engineering/genetic testing strategies Not started yet 6.6 WP5- Discovery of common divergent pathways for dormant stages, dormancy and desiccation resistance. Not started yet. 6.7 WP6 – Dissemination of results. 6.7.1 A workshop on dormancy. Place and time Berlin (MPI), May 2008 Suggested titles Sleeping beauties: Dormancy in organism Molecular, proteomic and metabolomic aspects of dormancy Dormancy and desiccation: molecular, proteomic and metabolomic aspects Genes, proteins and compounds associated with dormancy Dormancy: genes proteins and compound Suggested sessions Dormancy - mechanisms associated with onset of dormancy Mechanisms associated with “revival” (germination; hatching; renewal of metabolism) 10 Hibernation Signal transduction Compound in dormant forms Duration and agenda Duration of the workshop should be 2-3 days. The proposed agenda for each day is: Day1 Day 2 Day 3 Session 1: 09:00-10:30 Session 2: 11:00-12:30 Session 3: 14:00-15:30 Session 4: 16:00-18:00 (including round table discussion) Session 5: 09:00-10:30 Session 6: 11:00-12:30 Session 7: Posters? Session 8: 16:00-18:00 (including round table discussion) Session 9: 09:00-10:30 Session 10: 11:00-12:30 Session 11: 14:00-15:30 Session 12: 16:00-18:00 Festive Dinner Day 3 Total: 18 hours (36-45 oral presentations) Invited speakers:~12 (~10,000 Euro) or 24 (~20,000 Euro) Students :6,000 Euro Addition expenses: ~5,000 Euro Total: ~23,000 or 35,000 Euro Scientific organizing committee Chairpersons are to be decided. Proposed persons for the organizing: Microorganisms: Stefan? ,W. Hess? Plants: Derek Bewley, Henk Hillhorst Algae and macroalgae : Catherine Boyen? Invertebrates: Clegg? Tunnacliffe? Esther? 11 Insects: Roger? Vertebrates: Ken Storey? Joan? Compounds? Each session will be organized by 1 person from SB with a leading scientist in the field. Funding The workshop is partially budgeted by the project. It is suggested to apply to EMBO for additional funding. The deadline for submitting the proposal is February, 2007. The proposal should be submitted by one of the European partners and not by Esther because Israel is not a member of EMBO. 6.7.2 Publication of a book on dormancy Stefan will take care for publishing the book in a series that he is editing. The process will take about 2 years. It is suggested that people who will participate in the workshop will be invited to write reviews in the book. 7 Issues to be discussed in the next meeting Partners are requested to send the abstract of their report to Esther a month before the meeting. 7.1 ESTs and micorarrays Partners requests for additional ESTs and microarray slided. The cost of additional ESTs and microarray experiments will be supplied by Richard. Workshop on microarray experiments. Workshop on microarray analysis. 7.2 Workshop on dormancy Title Sessions Participants Organizing committee 12