B.3.1. J.F. Aldana Montes, R. Berlanga
advertisement

A Pilot Study Experience on Generating
Complex Ontology Instances from
Scientific Bibliographies on Real
Biological Domains
José F. Aldana-Montes1, Rafael Berlanga-Llavorí2, Roxana Danger2, Raul MontañésMartínez3, Mª del Mar Rojano-Muñoz1, Francisca Sánchez-Jiménez3
1
Khaos Research Group. Dpt. of Computer Languages and Computing Science,
University of Malaga, Campus de Teatinos. 29071 Malaga, Spain
http://Khaos.uma.es, E-mail: jfam@lcc.uma.es
2 Temporal
Knowledge Bases Group. Dpt. of Computer Languages and Computer
Systems, University Jaume I, Castellón, Spain
http://www3.uji.es/~berlanga/; E-mail: {berlanga, Roxana.Danger}@uji.es
3
ProCel Lab. Department of Molecular Biology and Biochemistry. Faculty of
Sciences, University of Málaga, Campus de Teatinos. 29071 Malaga, Spain
http://www.bmbq.uma.es/procel, E-mail: kika@uma.es
Abstract
In this paper we present a first insight into the generation of complex ontology instances
from scientific bibliographies like the one in PubMed/PubChem on a real biological
domain: Polyamines and Histamine. There is evidence for the involvement of both in
cancer and other inflammation- and/or angiogenesis-dependent diseases, but multiple
questions concerning the molecular processes behind these effects still remain to be
solved.
We address the problem of automatically generating ontology instances starting from a
collection of PDF documents stored in a bibliographic database. We have developed a
domain ontology which models and describes what we are searching for. The structure
of the document is extracted in order to generate a mapping between the ontology and
the document text. Using this mapping the ontology is populated with the extracted
knowledge.
1. Introduction
The Semantic Web (SW) is intended to enrich the current Web by creating
knowledge objects that allow both users and programs to better exploit the Web
resources [1]. The cornerstone of the Semantic Web is the definition of an ontology
that conceptualizes the knowledge within the resources of a particular domain [2].
Thus, the contents of the Web will be described in the future by means of a large
collection of semantically tagged resources that must be reliable and meaningful.
Nowadays most of the information on the Web consists of text documents with little
or no structure at all, which makes their manual annotation impracticable [3].
In automatic document classification several methods have been proposed to
automatically classify documents according to a given taxonomy of concepts (e.g.
[4]). However, these methods require a large amount of training data, and they do
not generate instances of the ontology. In the Information Extraction (IE) area, much
work has been focused on recognizing predefined entities (e.g. dates, locations,
names, etc.), as well as extracting relevant relations between them by using natural
language processing. However, current IE systems are restricted to extracting flat
and very simple relations, mainly to feed a relational database. Ontology
relationships are more complex as they can involve nested concepts and they have
associated inference rules.
For these reasons we address the problem of annotating texts according to an
ontology as a mapping problem between text fragments and ontology subgraphs. As
in IE systems, we also recognize named entities (e.g. protein names) that can be
directly associated to ontology concepts. However, unlike IE systems, we do not use
either syntactic or semantic analysis to extract the relations appearing in the text. As
a result, we can extract complex instances efficiently and effectively.
In this paper, we will apply this technique to extract instances from the biogenic
amines domain and all the related processes.
2. Generation of ontology instances
In this section we introduce the main formal aspects of the proposed extraction
technique presented in [2]. We adopt the formal definition of ontology from [6]. This
definition distinguishes between the conceptual schema of the ontology, which
consists of a set of concepts and their relationships, and its associated resource
descriptions, which are called instances. In our approach, we represent both parts
as oriented graphs, over which the ontology inference rules are applied to extract
and validate complex instances.
Table 1 presents the definitions of the concepts used when generating partial and
complete instances from words and entities appearing within a text fragment.
Definition
Formal description
Description
Def. 1.
- The set of the ontology concepts C is the subset of nodes in N that are not pointed by any arc
labeled as “instance_of”.
An ontology is a labeled oriented
graph G = (N, A) where the set of
nodes N contains both the concept
names and the instance identifiers,
and A is a set of labeled arcs
between them representing their
relationships.
-The taxonomy of concepts C consists of the subgraph that only contains the is_a arcs. It defines a
partial order over the elements of C, denoted with C.
-The relation signature is a function : R C x C, which contains the subsets of arc labels that
involve only concepts.
-The function dom: R C with dom(r) = 1((r)) gives the domain of a relation rR, whereas
range: R C with range(r) = 2((r)) gives its range.
-The function domC: C 2 gives the domain of definition of a concept.
Additionally, over the graph we
introduce the following restrictions
according to [6]:
-The function in order to express the proximity of two concepts c and c’ where dT(c, c’) and dR(c,
c’) are the distances between the two concepts c and c’.
Rel (c, c' ) 1- log C min {d T (c,c' ), d R (c,c' ) }
Def. 2.
We denote with
I
c
o
an instance related to the object o of the class c or simply an instance, as the
subgraph of the ontology that relates the node o with any other, and there exists an arc (o, instance_of,
c) A.
I o { (o, r , o' ) /(o, instance_of, c ) A, r R, (r ) (c * , c' ),
c
o domc (c * ), o' domc (c' ), c * C c, r ' , c * C dom(r ' ) C c}
Notice that the arc (o, instance_of,
c) is not included in the instance
subgraph. An instance is empty if
the object o only participates in the
arc (o, instance_of, c), which is
represented as the set {(o, *, *)}
Def. 3.
c
I
We call a specialization of the instance
o
c c '
I o o' , the following
to the class c’, ccc’, denoted with
instance:
c'
c
I o ' { (o' , r , x) /(o, r , x) I o , r ' R, r R r ' , dom(r ) C c' , range(r ) cx ,
.
represent an overridden property
x domC (cx ), o' domC (c' )}
Def. 4.
I
We call an abstraction of the instance
c'
c
o
Similarly to the previous definition,
we can obtain an abstract instance
by selecting all the triples whose
relation name can be abstracted to a
relation of the target class c’ and
renaming accordingly the instance
object.
cc'
to the class c’, c’cc, denoted with
I o o ' , the instance
c
I o' {(o' , r, x) /(o, r, x) I o , r, cx , dom(r ) C c' , range(r ) cx , x dom(cx )}
Def. 5:
c
c'
c
We call union of the instances I1 and I 2 and denote it by I1
o'
o
o
c
ˆ I2
I1 o
c'
o'
I c I
1 coc ' 2
I 1 o o '
c
I1 o
c'
I 2 o'
c ' c
I 2 o 'o
c'
Def. 6:
We call difference of the instances
c
I 1 o ˆ I 2
c'
o'
c
c
c 'c
I I
1 oc 2 oc''o c
I 1 o I 2 o 'o
c
I1 o
ˆ I 2 c ' the set:
o'
I 1 o {( o,*,*)} I 2
if
o'
c'
c
o'
(o' ,*,*) (c c c' c' c)
I2
if
I2
if
I 1 o {( o,*,*)} c c c'
if
I 1 o {( o' ,*,*)} c' c c
and
c'
, denoted with
o'
I2
c c c'
if
c' c c
We call symmetric difference of the instances
{( o' ,*,*)} c' c c
c
if
Def. 7:
o'
o'
c
Definitions 5-7 are necessary to
define
the
unification
and
aggregation of simple instances
into complex ones.
{( o' ,*,*)} c c c'
if
c'
Basically, this definition says that
an instance can be specialized by
simply renaming the object with a
name from the target class in all the
instance triples that do not
c
c
c'
I1 o ˆ I 2 o'
, the following set:
c
c'
c'
I1 o and I 2 o ' , denoted with I1 o ˆ I 2 o' the result of
c
c'
ˆ I 2 c' ˆ I1 c .
I1 o ˆ I 2 o'
o'
o
Def. 8:
We say that two instances
c
I1 o
and
c'
I 2 o'
related to the objects o and o’ of classes c and c’,
respectively, c C c’, are complementary if they satisfy at least one of the following two conditions:
1.
c c'
relations
x x’, r is biyective) and
((o, r, x), (o’, r’, x’) I1
c'
c
ˆ I 2 , r R r’, r r’, x x’) or
o
o'
at least one of the instances
I1 o
c
2.
c'
I1 o o' I 2 o' ,
((o’, r, x), (o’, r, x’)
I 2 o'
is empty.
c'
c
say
c'
that
c c '
two
c 'c '
ˆ I2
I o I1 o o
o 'o
u
complementary
u
Def. 10:
We say that two instances,
.
c'
or
Def. 9:
We
Two instances are complementary
if contradictions do not exist
between the values of their similar
instances
I1 o
and
I 2 o' are
unifiable
in
, c C c’.
u
c
c'
I1 o and I 2 o'
c*
are aggregable in
c c *
I o I1 o o {(o, r , o' ' )} , if
rR, (r) = (c*, c’’), c C c*, c’’C c’, and o’’ is the name of the instance
c ''
c 'c ''
I o '' I 2 o 'o '' .
Tabla 1. Definitions of the extraction method taken from [2]
Taking into account the definitions above, the system generates the set of instances
that are associated to each text fragment as follows. Firstly, it constructs all the
possible partial instances with the concept, entity and relation names that appear in
the text fragment. Then it tries to unify partial instances, substituting them by the
unified ones. Finally, instances are aggregated to form complex instances. The
following algorithm sums up the process. Further details of this process are given in
[2].
3. Generating Complex Ontology Instances from Scientific Bibliographics on the
Domain of the Biogenic Amines
3.1 The Domain of the Biogenic Amines Metabolism
From a molecular point of view, Biology is an informational science with three major
types of interconnected information: i) the structure of genes, along with the threedimensional structures of proteins, ii) the molecular machines of life, and iii)
biological systems with their emergent behaviour. Validation of new
biocomputational tools for biological data mining and integration requires experience
on a wide body of biological knowledge, which is essential: i) to select a proper and
interesting problem, ii) to build the underlying ontology, iii) to purge any automatic
searching result, and to build new applications (i.e.: hypothesis and conclusions) on
the obtained data. In this project, we have tried to accomplish all of the above
mentioned requirements to build a tool for location of molecular information by text
mining in both web pages and PDF documents. Our group combines experts in
computer science and molecular biology, involved in interdisciplinary projects for the
last 4 years. As a pilot topic for validation, we chose a module of the secondary
metabolism of living cells "the biogenic amine metabolism".
Biogenic amines are derived from amino acids. Polyamines play a relevant
regulatory role in macromolecular synthesis and cell proliferation rates [7-9]. On the
other hand, histamine is a biogenic amine synthesized by the enzyme histidine
decarboxylase (HDC) with a major role in immune response, inflammation, and
other multiple physiological activities [10-11]. All together biogenic amines are
involved in many of the most important and general diseases, such as neurological
disorders, cancer, and immune pathologies. Traditionally, histamine and polyamines
have been the subject of much interest by many biomedical research groups without
much communication among them. However, they share similar chemical and
metabolic facts and physiopathological missions [8-11]. Nevertheless, multiple
questions concerning the molecular processes behind these effects remain to be
solved. This may be due to the great dispersion of available data in specialized
literature of very different areas of biomedicine. For all of these reasons, this field
seems to be worthy of selection as an interesting conceptual benchmark. In silico
predictive models have been developed and published by the group at different
levels that help to explain many of the previous experimental observations, giving
rise to new findings and hypothesis [10,12,13]. Development of new text mining
tools is essential for efficiency in the progression of these projects.
3.2 Domain Ontology
Enzymes (mainly proteins) are the catalysts of the biological reactions that take part
in any metabolic pathway. The half-life of a given enzyme is determined by the
balance between its synthesis and degradation rates. Protein degradation involves
many different mechanisms (proteinase activities) specific for each enzyme. Many of
the regulatory mechanisms of amine metabolic pathways involved enzyme
destruction after the biological mission of their respective amine has been carried
out, as a mechanism to ensure an effective switch-off of the process. In addition,
several amine-related enzymes are synthetized in an unactive form that needs to be
cleavaged by proteinases to reach its active stage (processing /maturation). This is
the case for mammalian histidine decarboxylase, a very unstable enzyme that
requires a proteolytic mechanism for both maturation and regulation of its turnover.
A lot of information on the enzyme degradation/processing mechanisms involved in
amine metabolism has been generated in different text formats. An important
percentage of this information has been generated by collaborator groups and our
own group, especially in the case of mammalian histidine decarboxylase [14-17].
This seems to be a good test bank for our pilot project on text mining. Thus, we
adopted HDC as the pilot molecule to contrast our data mining efforts to develop an
automatized searching tool on published pdfs and other document formats.
Nevertheless, due to the highly consistent nature of protein metabolism, once the
data mining process is validated on this enzyme, it could be easily scaled-up to any
other enzyme, related or not to amine metabolism. This ontology has been
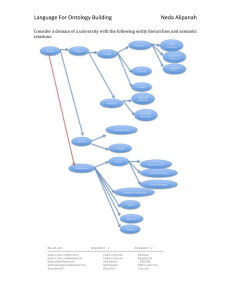
developed in OWL [18].
Fig. 2. Ontology
3.3 Benchmark
The benchmark has been chosen from an extensive pool of scientific works coming
from a world-wide group of experts on degradation and maturation of histidine
decarboxylase (HDC). In this benchmark 67 documents are compiled in PDF format
and eight web pages, 50 percent of them conform with the pertinent information
(highly-related to HDC degradation/maturation). Both collateral (related to other facts
of histamine metabolism) and unrelated information which were included as controls,
are essential for a correct and accurate validation of both the analyzers and the
ontology during this pilot experience. Document selection was done on the basis of
the impact and availability of the publication source. Selected documents content
dispersed (and in some cases contradictory) experimental results developed in the
field of the molecular biology and biochemistry.
Text description and figure of instances
<Sample id="004" source="22.pdf" section="Title and Abstract">
<P>Expression of 74-kDa histidine decarboxylase protein in a macrophage-like
cell line RAW 264.7 and inhibition by dexamethasone</P>
Total words
Instances
Correct Instances
Failed Instances
17
5
4
1
<Sample id="010" source="9.pdf" section="2.1.1">
<P>
The human H1 receptor contains 486, 488 or 487 amino acids in rat, mouse and
human respectively. It shares the typical features of GPCR, namely: seven
transmembrane domains of 20-25 amino-acids predicted to form an a-helice that
spans the plasma membrane and an extracellular NH2 terminal (polypeptide)
domain with glycosylation site. It is encoded by a single exon gene located on
the distal short arm of chromosome 3p25 in humans and chromosome 6 in mice.
Histamine binds to aspartate residues in the transmembrane domain (motif) 3 of
the receptor and to asparagine + lysine residues within the transmembrane
domain 5. H1 receptors are involved in the pathological process of allergy such
as allergenic rhinitis, conjunctivitis, atopic dermatitis, urticaria, asthma
and anaphylaxis. In the lung, it mediates bronchoconstriction and increased
vascular permeability. The H1R is expressed in numerous cell types, including
airway and vascular smooth muscle cells, hepatocytes, chondrocytes, nerve
cells, endothelial cells, neutrophils, monocytes, dendritic cells, as well as T
and B lymphocytes, in which it mediates the various biological manifestations
of allergic responses [3-5]. H1R is a Gaq/11-coupled protein (polypeptide)
with a very large third intracellular loop and a relatively short C-terminal
(polypeptide) tail (Fig. 1). The main signal induced by ligand binding is the
activation of phospholipase C-generating inositol 1,4,5-triphosphate (Ins
(1,4,5) P3) and 1,2-diacylglycerol leading to increased cytosolic (cellular
lication) Ca2+ [4]. This rise in intra-cellular calcium levels seems to account
for the various pharmacological activities promoted by the receptor, such as
nitric oxide production, vasodilatation, liberation of arachidonic acid from
phospholipids and increased cyclic AMP.H1R also activates NF kB through Gaq11
and Gbg upon agonist binding, while constitutive activation of NF kB occurs
through Gbg only [6].
</P>
Total words
Instances
Correct Instances
Failed Instances
283
14
10
4
Table 2. Representation of total correct instances (orange diamond in figure) of text and the fails
instances (gray diamond).
Concepts, links and instances were defined on the basis of scientific criteria without
taking into account the precise benchmark-containing information or semantic style.
One of the major difficulties detected throughout the work was the semantic
dispersion in the expression of concepts and results in the biochemical information,
which make the accurate definition of the concepts and instances for the mining
difficult, and obliged to the establishment of an enriched list of synonyms. Working
in this proving bank we obtain a set of results as shown in the examples in table 2.
The results indicate that concepts contained in the text are properly detected;
although the system fails to detect the appropriate context, the results are not
discouraging at all; however, further efforts will be necessary to restrict possibilities
to establish long-distance relationships (with respect to the text position of the
concepts) and to solve problems with ambiguity of the biological terms (for instance,
in literature, H1 could mean location of a histidine residue at position 1 as well as
the abbreviation for a histamine receptor sybtype). These changes would lead to
increase the astringency of the search criteria, and consequently to the loss of some
information, although accuracy should be improved
The extraction algorithm captures the main relationships between correctly detected
entities, but it mainly fails in the correct identification of some basic entities. This is
principally because we have used preliminary regular expressions to detect named
entities instead of a good thesaurus. Another problem we identify is the correct
tokenization of text fragments due to the large number of chemical formula and
abbreviations that are included in biomedical texts. Future work must then focus on
improving the most basic mechanisms for detecting named entities and concept
boundaries in the texts. Finally, we have also noticed that the level of detail of the
ontology also affects the quality of extracted instances. Thus, the more detailed the
ontology is, the likelier and better are the relations found in text fragments. In this
respect, much more work must be done to improve the details of the target ontology.
4. Conclusion
We have presented a pilot project on the generation of complex ontology instances
in the very real (and thus hard) domain of the biogenic amines metabolism (BAM).
An ontology which modelizes the discussion domain of the BAM has been
developed in OWL. We have built a benchmark from the real bibliography of the
BAM domain (extracted from several public domain bibliographic databases) and
we are going to use this pilot experience to evaluate in a well-known controlled
environment the quality of the results produced by our system. Firsts results are not
absolutely discouraging but further work must be done in the description of the
ontology.
Acknowledgements
This work was supported by Grants CVI-267, CVI-657, TIN-09098-C05-01 and
TIN2005-09098-C05-04.
References
[1] Berners-Lee, T., Hendler, J., Lassila, O. "The Semantic Web". Scientific American, 2001.
[2] Danger, R., Sanz, I., Berlanga-Llavori, R., Ruiz-Shulcloper, J. “A Proposal for the Automatic
Generation of Instances from Unstructured Text”, Lecture Notes in Computer Science, Volume 3287,
pp. 462 – 469, 2004.
[3] Gruber, T.R. “Towards Principles for the Design of Ontologies used for Knowledge Sharing”,
International Journal of Human-Computer Studies Vol. 43, pp. 907-928, 1995.
[4] Forno, F., Farinetti, L., Mehan, S. “Can Data Mining Techniques Ease The Semantic Tagging
Burden?”, SWDB 2003, pp. 277-292, 2003.
[5] Doan, A. et al. “Learning to match ontologies on the Semantic Web”. VLDB Journal 12(4), pp. 303319, 2003.
[6] Maedche, A., Neumann, G. and Staab, S. “Bootstrapping an Ontology based Information Extraction
System”. Studies in Fuzziness and Soft Computing, Springer, 2001.
[7] Cohen SS. Aguide to polyamines. Oxford University Press, New York; 1998.
[8] Thomas T, Thomas TJ. Polyamines in cell growth and cell death: molecular mechanisms and
therapeutic applications. Cell Mol Life Sci. 2001; 58: 244–58.
[9] Medina MA, Urdiales JL, Rodriguez-Caso C, Ramirez FJ, Sanchez-Jimenez F. Biogenic amines and
polyamines: similar biochemistry for different physiologi cal missions and biomedical applications. Crit
Rev Biochem Mol Biol. 2003; 38: 23–59.
[10] Moya-Garcia AA, Medina MA, Sanchez-Jimenez F. Mammalian histidine decarboxylase: from
structure to function. Bioessays 2005; 27: 57–63.
[11] Medina MA, Quesada AR, Nunez de Castro I, Sanchez-Jimenez F. Histamine, polyamines, and
cancer. Biochem Pharmacol. 1999; 57: 1341–4.
[12] Medina M.A. Correa-Fiz F., Rodríguez-Caso C., Sánchez-Jiménez F. A comprehensive view of
polyamine and histamine metabolism to the light of new technologies. J. Cell. Mol. Med. 2005. Vol. 9.
854-864.
[13] Rodríguez-Caso C., Montañez R., Cascante M., Sánchez-Jiménez F., Medina M.A. Mathematical
modeling of polyamine metabolism in mammals. J. Biol. Chem. en prensa (Mayo,06), on-line PMID:
16709566.
[14] Ángel N., Olmo M.T., Coleman C.S., Medina M. A., Pegg A.E., Sánchez-Jiménez F. Experimental
evidence for structure/activity features in common between mammalian. Biochem. J. 1996. Vol. 320.
365-368.
[15] Matés J.M., del Valle A.E., Urdiales J.L., Coleman C.S., Feith D., Pegg A.E., Sánchez-Jiménez
F.Structure/function relationship studies on the T/S residues 173-177 of rat ODC. Biochem. Biophys.
Acta 1998. Vol. 1386. 113-120.
[16] Olmo M.T. Urdiales J.L., Pegg A.E., Medina M.A., Sánchez-Jiménez F.. Eur. J. Biochem. 2000. Vol.
267. 1527-1531.
[17] Olmo M.T., Sánchez-Jiménez F., Medina M.A., Hayashi H. Spectroscopic análisis of recombinant rat
histidina decarboxylase. 2002. Vol.132. 433-439.
[18] OWL Web Ontology Language 1.0 Reference. W3C Candidate Recommendation 15 Decembre 2003
http://www.w3.org/TR/owl-ref.