University of Göttingen
advertisement

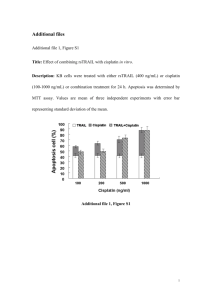
Section 15.5 - Assays for Apoptosis. Molecular Probes Chapter Apoptosis (programmed cell death) is the genetically controlled ablation of cells during normal development. Inappropriately regulated apoptosis is implicated in disease states such as Alzheimer's disease, stroke and cancer. Apoptosis is distinct from necrosis in both the biochemical and the morphological changes that occur. In contrast to necrotic cells, apoptotic cells are characterized morphologically by compaction of the nuclear chromatin, shrinkage of the cytoplasm and production of membrane-bound apoptotic bodies. Biochemically, apoptosis is distinguished by fragmentation of the genome and cleavage or degradation of several cellular proteins. As with cell viability, no single parameter fully defines cell death in all systems; therefore, it is often advantageous to use several different approaches when studying apoptosis. Several methods have been developed to distinguish live cells from early and late apoptotic cells and from necrotic cells; these are described below and in a number of review articles and seminal publications. Anti-cancer drug candidates failing to induce apoptosis are likely to have decreased clinical efficacy, making apoptosis assays important tools for high-throughput drug screening. Apoptotic cells are typically eliminated by phagocytosis; thus, apoptotic cells that have been selectively labeled with a fluorescent dye can potentially be used as tracers for phagocytosis, a cell process that is discussed in Section 16.1. Apoptosis Assays Using Nucleic Acid Stains DNA Stains For Detection of Apoptotic Cells The characteristic breakdown of the nucleus during apoptosis comprises collapse and fragmentation of the chromatin, degradation of the nuclear envelope and nuclear blebbing, resulting in the formation of micronuclei. Therefore, nucleic acid stains can be useful tools for identifying even low numbers of apoptotic cells in cell populations. Several nucleic acid stains, all of which are listed in Section 8.1, have been used to detect apoptotic cells by fluorescence imaging or flow cytometry. * Our YO-PRO-1 (Y3603) nucleic acid stain is the basis of an important assay for apoptotic cells that is compatible with both fluorescence microscopy and flow cytometry. Selective uptake of YO-PRO-1 by apoptotic cells of a dexamethasone-treated population of thymocytes, an irradiated peripheral blood mononuclear cell population and a growth factor–depleted tumor B cell line was confirmed by cell sorting. Unlike Hoechst 33342 staining, YO-PRO-1 staining had no effect on the ability of stained T cells to proliferate. Moreover, the visible-light absorption of the YO-PRO-1 stain () eliminates the need for UV excitation capabilities in flow cytometry. YO-PRO-1 is the key reagent in our Vybrant Apoptosis Assay Kits #4 and #7 (V13243, V23201, see below), which provide the reagents and tested protocols for combination flow cytometric apoptosis and necrosis assays. * Some of our cell-permeant, green-fluorescent SYTO dyes, including the SYTO 13 and SYTO 16 nucleic acid stains (S7575, S7578), are proving useful for distinguishing apoptotic neuronal cells and apoptotic thymocytes. Our SYTO Fluorescent Nucleic Acid Stain Sampler Kits (S7554, S7572, S11340, S11350, S11360; Section 8.1) provide fluorescent SYTO dyes covering the entire visible spectrum (Table 8.3) that may be screened for their utility in monitoring apoptosis. In addition, apoptotic cells in a follicular lymphoma cell line could be discriminated earlier with our SYTO 17 red-fluorescent nucleic acid stain (S7579) than with either fluorescein-labeled annexin V or propidium iodide. * Hoechst 33342 (H1399, H3570; FluoroPure Grade, H21492) is readily taken up by cells during the initial stages of apoptosis, whereas cell-impermeant dyes such as propidium iodide (P1304MP, P3566, P21493; Section 8.1) and ethidium bromide (E1305, E3565; Section 8.1) are excluded. Later stages of apoptosis are accompanied by an increase in membrane permeability, which allows propidium iodide to enter cells. Thus, a combination of Hoechst 33342 and propidium iodide has been extensively used for simultaneous flow cytometric and fluorescence imaging analysis of the stages of apoptosis and cell-cycle distribution. Our Vybrant Apoptosis Assay Kit #5 (V13244, see below) is based on these reagents and our Vybrant Apoptosis Assay Kit #7 (V23201, see below) adds the YO-PRO-1 nucleic acid to selectively determine the apoptotic cell population in a three-color experiment. * The rate of Hoechst 33342 uptake in partially apoptotic cell populations is correlated with low intracellular pH, as measured with our carboxy SNARF-1 pH indicator (C1271, C1272; Section 21.2). * Hoechst 33342, which selectively stains nuclei of apoptotic cells blue fluorescent, has also been used in combination with calcein AM (C1430, C3099, C3100MP; Section 15.2), which stains all cells that have intact membranes — even apoptotic cells — green fluorescent. Presumably the dead-cell population could be selectively detected using propidium iodide to make this a three-color assay. * 7-Aminoactinomycin D (7-AAD, A1310) has been used alone or in combination with Hoechst 33342 to separate populations of live cells, early apoptotic cells and late apoptotic cells by flow cytometry. The staining pattern of 7-AAD is retained following cell fixation, and its unusually large Stokes shift is advantageous when simultaneously staining with cell-surface labels. 7-AAD staining has also been used to detect apoptotic cells by their characteristic morphology using fluorescence microscopy. 7-AAD has also been used in combination with the green-fluorescent SYTO 16 nucleic acid stain (S7578) to detect early stages of apoptosis that could not be detected by 7-AAD alone. * The cell-permeant nucleic acid stain LDS 751 (L7595) has been used to discriminate intact nucleated cells from nonnucleated cells and cells with damaged nuclei, as well as to differentiate apoptotic cells from nonapoptotic cells. * Acridine orange (A1301, A3568) exhibits metachromatic fluorescence that is sensitive to DNA conformation, making it a useful probe for detecting apoptotic cells. When analyzed by flow cytometry, apoptotic cells stained by acridine orange show reduced green fluorescence and enhanced red fluorescence in comparison to normal cells. * DAPI (D1306, D21490; Section 8.1) and sulforhodamine 101 (S359, Section 14.3) can be used together in fixed apoptotic cells to reveal concomitant breakdown of proteins and DNA. * The excited-state lifetime of ethidium homodimer-2 (E3599, Section 8.1) has been shown to be different in populations of aldehyde-fixed apoptotic and nonapoptotic cells. * Ethidium monoazide (E1374, Section 15.2) passes through the partially compromised membrane of apoptotic cells; photolysis results in covalent labeling of intracellular nucleic acids that persists through fixation and permeabilization. DNA fragmentation can also be detected in vitro using electrophoresis. DNA extracted from apoptotic cells, separated by gel electrophoresis and stained with ethidium bromide reveals a characteristic ladder pattern of low molecular weight DNA fragments. Ethidium bromide has been used for a dot-blot assay to detect apoptotic DNA fragments. Our ultrasensitive SYBR Green I nucleic acid stain (S7567, Section 8.4) and SYBR DX DNA blot stain (S7550, Section 8.5) allow the detection of even fewer apoptotic cells in these applications (). Electrophoresis of apoptotic cells in an agarose gel matrix results in the formation of distinctive "comets" of DNA leaking from apoptotic cells (but not normal cells; see the paragraph, Comet (Single-Cell Gel Electrophoresis) Assay to Detect Damaged DNA, below) (). Vybrant Apoptosis Assay Kit #4 Our Vybrant Apoptosis Assay Kit #4 (V13243) detects apoptosis on the basis of changes that occur in the permeability of cell membranes (Table 15.4). This kit contains ready-to-use solutions of both the YO-PRO-1 and propidium iodide nucleic acid stains. Our Patented YO-PRO-1 nucleic acid stain selectively passes through the plasma membranes of apoptotic cells and labels them with moderate green fluorescence. Necrotic cells are stained with the red-fluorescent propidium iodide, a DNA-selective dye that is membrane impermeant but that easily passes through the compromised plasma membranes of necrotic cells. Live cells are not appreciably stained by either YO-PRO-1 or propidium iodide. The dyes included in the Vybrant Apoptosis Assay Kit #4 are effectively excited by the 488 nm spectral line of the argon-ion laser and are useful for both flow cytometry (Figure 15.80) and fluorescence microscopy (). We have optimized Our Vybrant Apoptosis Assay Kits using Jurkat cells, a human T-cell leukemia clone, treated with camptothecin to induce apoptosis. Some modifications may be required for use with other cell types. The kit components, number of assays and assay principles are summarized in Table 15.4. Vybrant Apoptosis Assay Kits #5 and #7 The Vybrant Apoptosis Assay Kit #5 (V13244) provides a rapid and convenient assay for apoptosis based upon fluorescence detection of the compacted state of the chromatin in apoptotic cells. This kit contains ready-to-use solutions of the blue-fluorescent Hoechst 33342 dye (excitation/emission maxima ~350/461 nm when bound to DNA), which stains the condensed chromatin of apoptotic cells more brightly than the chromatin of nonapoptotic cells, and the red-fluorescent propidium iodide (excitation/emission maxima ~535/617 nm when bound to DNA), which is permeant only to dead cells with compromised membranes (Table 15.4). The staining pattern resulting from the simultaneous use of these dyes makes it possible to distinguish normal, apoptotic and dead cell populations by flow cytometry or fluorescence microscopy. The 351 nm spectral line of an argon-ion laser or other suitable UV source is required for excitation of the Hoechst 33342 dye, whereas propidium iodide may be excited with the 488 nm spectral line of an argon-ion laser. We have optimized this assay using Jurkat cells, a human T-cell leukemia clone, treated with camptothecin to induce apoptosis. Some modifications may be required for use with other cell types. The kit components, number of assays and assay principles are summarized in Table 15.4. The Vybrant Apoptosis Assay Kit #7 combines the detection principles used in our Vybrant Apoptosis Assay Kits #4 (see above) and #5. Three nucleic acid stains — Hoechst 33342, YO-PRO-1 and propidium iodide — are utilized to identify by flow cytometry the fully live-cell population by their blue fluorescence, the greenfluorescent apoptotic population and the red-fluorescent dead-cell population. The stains are provided as separate solutions to facilitate optimization of the assay for the cell line under study and the equipment available. However, once optimized, the assay can be completed using simultaneous staining with a mixture of the three nucleic acid stains and either UV excitation of all three dyes or with a combination of UV excitation for the Hoechst 33342 dye and excitation by the 488 nm spectral line of the argon-ion laser. Differences in the intensity of the dye staining may make it difficult to simultaneously photograph the live, apoptotic and dead cells by microscopy. The kit components, number of assays and assay principles are summarized in Table 15.4. Comet (Single-Cell Gel Electrophoresis) Assay to Detect Damaged DNA The Comet assay, or single-cell gel electrophoresis assay, is used for rapid detection and quantitation of DNA damage from single cells. The Comet assay is based on the alkaline lysis of labile DNA at sites of damage. Cells are immobilized in a thin agarose matrix on slides and gently lysed. When subjected to electrophoresis, the unwound, relaxed DNA migrates out of the cells. After staining with a nucleic acid stain, cells that have accumulated DNA damage appear as fluorescent comets, with tails of DNA fragmentation or unwinding (). In contrast, cells with normal, undamaged DNA appear as round dots, because their intact DNA does not migrate out of the cell. The ease and sensitivity of the Comet assay has provided a fast and convenient way to measure damage to human sperm DNA, evaluate DNA replicative integrity, monitor the sensitivity of tumor cells to radiation damage and assess the sensitivity of molluscan cells to toxins in the environment. The Comet assay can also be used in combination with FISH (Section 8.5) to identify specific sequences with damaged DNA. Comet assays have traditionally been performed using ethidium bromide to stain the DNA. However, our YOYO-1 dye was found to increase the sensitivity of the assay eightfold compared to ethidium bromide. Use of the SYBR Gold and SYBR Green I stains improves the sensitivity of this assay (). Detecting DNA Strand Breaks with ChromaTide Nucleotides DNA fragmentation that occurs during apoptosis produces DNA strand breaks. TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) assays are widely used for detecting DNA nicks in apoptotic cells. Once the cells are fixed, DNA strand breaks can be detected in situ using mammalian terminal deoxynucleotidyl transferase (TdT), which covalently adds labeled nucleotides to the 3'-hydroxyl ends of these DNA fragments in a template-independent fashion. Break sites have traditionally been labeled with biotinylated dUTP, followed by subsequent detection with an avidin or streptavidin conjugate (Section 7.6, Table 7.22). However, a more direct approach for detecting DNA strand breaks in apoptotic cells is possible via the use of our ChromaTide BODIPY FL-14-dUTP (C7614) as a TdT substrate (). The single-step BODIPY FL dye–based assay has several advantages over indirect detection of biotinylated or haptenylated nucleotides, including fewer protocol steps and increased cell yields. BODIPY FL dye– labeled nucleotides have also proven superior to fluorescein-labeled nucleotides for detection of DNA strand breaks in apoptotic cells because they provide stronger signals, a narrower emission spectrum and less photobleaching (). Moreover, it has been reported that BODIPY FL-14-dUTP incorporated into the granules of the condensed chromatin structure of late-apoptotic cells — cells characterized by extensive nuclear fragmentation — exhibits yellow fluorescence, whereas uncondensed areas of the nuclei or early-apoptotic cells exhibit green fluorescence. This spectral shift, which is characteristic of the BODIPY fluorophores, is most likely a consequence of stacking of the BODIPY FL fluorophores (Figure 13.6) and could be very useful for identifying the stages of apoptosis on a single-cell basis. Our Texas Red-12-dUTP (C7631) has been used similarly for a TdT-mediated apoptosis assay; presumably a number of the ChromaTide dUTP nucleotides listed in Table 8.7 could be used for the direct or indirect TUNEL assay; we have not yet tried the ChromaTide dCTP nucleotides in this assay. Furthermore, our anti-dye antibodies (Section 7.4) can amplify the signal of many of the dyes used to prepare the ChromaTide nucleotides. In situ DNA modifications by labeled nucleotides have been used to detect DNA fragmentation in what may be apoptotic cells in autopsy brains of Huntington's and Alzheimer's disease patients. DNA fragmentation is also associated with amyotrophic lateral sclerosis. Analogous to TdT's ability to label double-strand breaks, the E. coli repair enzyme DNA polymerase I can be used to detect single-strand nicks, which appear as a relatively early step in some apoptotic processes. Because our ChromaTide BODIPY FL-14-dUTP (C7614) and ChromaTide fluorescein-12-dUTP (C7604) are incorporated into DNA by E. coli DNA polymerase I, it is likely that they may also be effective for in situ labeling with the nick translation method. APO-BrdU TUNEL Assay Kit Because DNA fragmentation is one of the most reliable methods for detecting apoptosis, we have collaborated with Phoenix Flow Systems to offer the APO-BrdU TUNEL Assay Kit (A23210), which provides all the materials necessary to label and detect the DNA strand breaks of apoptotic cells. When DNA strands are cleaved or nicked by nucleases, a large number of 3'-hydroxyl ends are exposed. In the APO-BrdU assay, these ends are labeled with BrdUTP and terminal deoxynucleotidyl transferase (TdT) using the TUNEL technique described above. Once incorporated into the DNA, BrdU is detected using an Alexa Fluor 488 dye–labeled anti-BrdU monoclonal antibody (). This kit also provides propidium iodide for determining total cellular DNA content, as well as fixed control cells for assessing assay performance. The APO-BrdU TUNEL Assay Kit includes complete protocols for use in flow cytometry applications, though it may also be adapted for use with fluorescence microscopy. Each kit contains: * Terminal deoxynucleotidyl transferase (TdT), for catalyzing the addition of BrdUTP at the break sites * 5-Bromo-2'-deoxyuridine 5'-triphosphate (BrdUTP) * Alexa Fluor 488 dye–labeled anti-BrdU mouse monoclonal antibody PRB-1, for detecting BrdU labels * Propidium iodide/RNase staining buffer, for quantitating total cellular DNA * Reaction, wash and rinse buffers * Positive control cells (a fixed human lymphoma cell line) * Negative control cells (a fixed human lymphoma cell line) * Detailed protocols (APO-BrdU TUNEL Assay Kit) Sufficient reagents are provided for approximately 60 assays of 1 mL samples, each containing 1–2 106 cells/mL. Apoptosis Assays Using Annexin V Conjugates Annexin V Conjugates Molecular Probes is collaborating with Nexins Research BV — the original developer and patent holder of fluorescent phosphatidylserine-binding proteins — to provide what we feel are the best and brightest annexin V conjugates available. The human vascular anticoagulant annexin V is a 35–36 kilodalton, Ca2+-dependent phospholipid-binding protein that has a high affinity for phosphatidylserine (PS). In normal viable cells, PS is located on the cytoplasmic surface of the cell membrane. However, in apoptotic cells, PS is translocated from the inner to the outer leaflet of the plasma membrane, exposing PS to the external cellular environment where it can be detected by annexin V conjugates. In leukocyte apoptosis, PS on the outer surface of the cell marks the cell for recognition and phagocytosis by macrophages. Highly fluorescent annexin V conjugates provide quick and reliable detection methods for studying the externalization of phosphatidylserine, an indicator of intermediate stages of apoptosis. Nuclear fragmentation, mitochondrial membrane potential flux and caspase-3 activation apparently precede phosphatidylserine "flipping" during apoptosis, while permeability to propidium iodide and cytoskeletal collapse occur later. The difference in fluorescence intensity between apoptotic and nonapoptotic cells stained by our fluorescent annexin V conjugates, as measured by flow cytometry, is typically about 100-fold (Figure 15.85). Annexin V conjugates (Annexin V Conjugates for Apoptosis Detection) are very useful for flow cytometry, confocal or epifluorescence microscopy and, like antibody staining, can be used in combination with other dyes, including nucleic acid stains, to accurately assess mixed populations of apoptotic and nonapoptotic cells. Our annexin V conjugates are available as standalone reagents, each suitable for 50–100 flow cytomety assays or many more imaging assays, or in several variations of our Vybrant Apoptosis Assay Kits (Table 15.4). Our annexin V conjugates include: * Alexa Fluor 488 annexin V (A13201, ), a green-fluorescent conjugate (excitation/emission maxima ~495/519 nm) that has spectral characteristics similar to fluorescein conjugates, but exhibits fluorescence that is brighter, much more photostable and less pH dependent (Figure 1.51, Figure 1.9; , ). Alexa Fluor 488 annexin V is used in both our Vybrant Apoptosis Assays Kits #1 and #2 (V13240, V13241; see below), which contain all of the reagents and an easy-to-follow protocol for flow cytometric detection and quantitation of apoptotic cells. * Fluorescein (FITC) annexin V (A13199), a green-fluorescent conjugate that has been extensively used by a number of laboratories to detect apoptotic cells populations. Fluorescein annexin V is frequently used in combination with propidium iodide to detect necrotic cells, as in our Vybrant Apoptosis Assay Kit #3 (V13242, see below). * Oregon Green 488 annexin V (A13200), a green-fluorescent conjugate that is spectrally similar to the fluorescein annexin V conjugate but is brighter and more photostable (Figure 1.44). * R-phycoerythrin annexin V (A35111), a highly fluorescent phycobiliprotein conjugate with absorption maxima at 496 nm, 546 nm and 565 nm and an emission maximum at 578 nm. * Allophycocyanin annexin V (A35110), a highly fluorescent phycobiliprotein conjugate with absorption/emission maxima of 650/660 nm. * Alexa Fluor 568 annexin V (A13202), a red-orange–fluorescent annexin V conjugate (excitation/emission maxima ~578/603 nm) with exceptionally bright and photostable fluorescence. We have determined that this conjugate can be used for simultaneous staining with green-fluorescent probes, such as our greenfluorescent Alexa Fluor 488 anti–CD 4 conjugate (A21335, Section 7.5), for multiparametric experiments. * Alexa Fluor 594 annexin V (A13203), a red-fluorescent annexin V conjugate with spectra similar to those of Texas Red conjugates (excitation/emission maxima ~590/617 nm) that can be used with green-fluorescent probes for multiparameter experiments. The Alexa Fluor 594 conjugate is readily excited by the 568 nm spectral line used in many confocal laser-scanning microscopes and has fluorescence that is well separated from the emission of green-fluorescent probes. * Alexa Fluor 647 annexin V (A23204), which permits use of long-wavelength excitation sources for detection of apoptotic cells by either flow cytometry or microscopy. * Alexa Fluor 350 annexin V (A23202) can be excited in the ultraviolet and has bright-blue fluorescence. Alternatively, the reagents in our Vybrant Apoptosis Assay Kit #6 (V23200) can be used in this spectral region. * Biotin-X annexin V (A13204), which can be detected by any of our fluorescent avidin or streptavidin conjugates (Section 7.6), gives the researcher the ultimate in color selection for multiparametric experiments. Biotin-X annexin V also permits detection of apoptotic cells by electron microscopy and should permit separation of apoptotic cells with our Captivate ferrofluid streptavidin conjugate (C21476, Section 7.6). Vybrant Apoptosis Assay Kit #1 With the Vybrant Apoptosis Assay Kit #1 (V13240), apoptotic cells are detected on the basis of the externalization of phosphatidylserine. This kit contains recombinant annexin V conjugated to the Alexa Fluor 488 dye, one of our brightest and most photostable green fluorophores to provide maximum sensitivity. In addition, the kit includes a ready-to-use solution of the SYTOX Green nucleic acid stain. The SYTOX Green dye is impermeant to live cells and apoptotic cells but stains necrotic cells with intense green fluorescence by binding to cellular nucleic acids. After staining a cell population with Alexa Fluor 488 annexin V and SYTOX Green dye in the provided binding buffer, apoptotic cells show green fluorescence, dead cells show a higher level of green fluorescence and live cells show little or no fluorescence (Figure 15.87). These populations can easily be distinguished using a flow cytometer with the 488 nm spectral line of an argon-ion laser for excitation. Both Alexa Fluor 488 annexin and the SYTOX Green dye emit a green fluorescence that can be detected in the FL1 channel, freeing the other channels for the detection of additional probes in multicolor labeling experiments. We have optimized Our Vybrant Apoptosis Assay Kits using Jurkat cells, a human Tcell leukemia clone, treated with camptothecin to induce apoptosis. Some modifications may be required for use with other cell types. The kit components, number of assays and assay principles are summarized in Table 15.4. Vybrant Apoptosis Assay Kit #2 Like the Vybrant Apoptosis Kit #1, our Vybrant Apoptosis Assay Kit #2 (V13241) detects the externalization of phosphatidylserine in apoptotic cells (Table 15.4). The Vybrant Apoptosis Assay Kit #2 provides a sensitive two-color assay that employs our green-fluorescent Alexa Fluor 488 annexin and a ready-to-use solution of the red-fluorescent propidium iodide nucleic acid stain. Propidium iodide is impermeant to live cells and apoptotic cells but stains necrotic cells with red fluorescence, binding tightly to the nucleic acids in the cell. After staining a cell population with Alexa Fluor 488 annexin V and propidium iodide in the provided binding buffer, apoptotic cells show green fluorescence, dead cells show red and green fluorescence, and live cells show little or no fluorescence (Figure 15.85). These populations can easily be distinguished using a flow cytometer with the 488 nm spectral line of an argon-ion laser for excitation. We have optimized this assay using Jurkat cells, a human T-cell leukemia clone, treated with camptothecin to induce apoptosis. Some modifications may be required for use with other cell types. The Vybrant Apoptosis Assay Kit #2 is designed for use with either flow cytometers or fluorescence microscopes. The kit components, number of assays and assay principles are summarized in Table 15.4. Vybrant Apoptosis Assay Kit #3 The Vybrant Apoptosis Assay Kit #3 (V13242) is very similar to the Vybrant Apoptosis Assay Kit #2, except that it contains fluorescein (FITC) annexin V in place of the Alexa Fluor 488 conjugate found in Kit #2 (Table 15.4). The kit components, number of assays and assay principles are summarized in Table 15.4. Vybrant Apoptosis Assay Kit #6 The Vybrant Apoptosis Assay Kit #6 (V23200) is very similar to the Vybrant Apoptosis Assay Kit #2, except that it contains biotin-X annexin V and Alexa Fluor 350 streptavidin in place of the Alexa Fluor 488 conjugate found in Kit #2 (Table 15.4). After staining a cell population with biotin-X annexin V in the provided binding buffer, Alexa Fluor 350 streptavidin is added to fluorescently label the bound annexin V. Finally, propidium iodide is added to detect necrotic cells. Apoptotic cells show blue fluorescence, dead cells show red and blue fluorescence and live cells show little or no fluorescence. These populations can easily be distinguished using a flow cytometer with UV excitation for the Alexa Fluor 350 fluorophore and 488 nm excitation for the propidium iodide. With the Vybrant Apoptosis Assay Kit #6, fluorescence in the green channel (FL1) is minimal. In the same experiment for apoptosis detection, the researcher can apply a green-fluorescent probe, for example an antibody labeled with the Alexa Fluor 488 dye or with fluorescein. The kit components, number of assays and assay principles are summarized in Table 15.4. Vybrant Apoptosis Assay Kit #8 The Vybrant Apoptosis Assay Kit #8 (V35112 is very similar to the Vybrant Apoptosis Assay Kit #1, except that it contains R-phycoerythrin (R-PE) annexin V instead of Alexa Fluor 488 annexin V. R-PE is an extremely fluorescent phycobiliprotein that can easily be excited with the 488 nm spectral line of the argonion laser on a standard flow cytometer and exhibits an emission maximum at 578 nm. In addition to R-PE annexin V, this kit includes the SYTOX Green nucleic acid stain, which is impermeant to live cells and apoptotic cells but stains necrotic cells with intense green fluorescence. After staining a cell population with R-PE annexin V and SYTOX Green stain, apoptotic cells show orange fluorescence with very little green fluorescence, late apoptotic cells show a higher level of green and orange fluorescence and live cells show little or no fluorescence (Figure 15.88). These populations can easily be distinguished using a flow cytometer with the 488 nm spectral line of an argon-ion laser for excitation. The kit components, number of assays and assay principles are summarized in Table 15.4. Vybrant Apoptosis Assay Kit #9 The Vybrant Apoptosis Assay Kit #9 (V35113 is very similar to the Vybrant Apoptosis Assay Kit #1, except that it contains allophycocyanin annexin V instead of Alexa Fluor 488 annexin V. Allophycocyanin is an extremely fluorescent phycobiliprotein that can easily be excited with the 633 nm spectral line of the He–Ne laser on a standard flow cytometer and exhibits an emission maximum at 660 nm. In addition to allophycocyanin annexin V, this kit includes the SYTOX Green nucleic acid stain, which is impermeant to live cells and apoptotic cells but stains necrotic cells with intense green fluorescence. After staining a cell population with allophycocyanin annexin V and SYTOX Green stain, apoptotic cells show far-red fluorescence with very little green fluorescence, late apoptotic cells show a higher level of green and far-red fluorescence and live cells show little or no fluorescence (Figure 15.89). These populations can easily be distinguished using a flow cytometer with both the 488 nm spectral line of an argon-ion laser and the 633 nm spectral line of a He–Ne laser for excitation. The kit components, number of assays and assay principles are summarized in Table 15.4. Vybrant Apoptosis Assay Kit #10 The Vybrant Apoptosis Assay Kit #10 (V35114 is very similar to the Vybrant Apoptosis Assay Kit #9, except that it also contains C12-resazurin. Nonfluorescent C12 resazurin is reduced by viable cells to orangefluorescent C12 resorufin. Resazurin has been used extensively to detect the metabolic activity of many different cell types, from bacteria to higher eukaryotes. After staining a cell population with allophycocyanin annexin V, C12-resazurin and SYTOX Green stain, apoptotic cells show far-red fluorescence, intermediate orange fluorescence and no green fluorescence; late apoptotic cells show intense far-red and green fluorescence and little orange fluorescence; live cells show little or no green or far-red fluorescence but significant orange fluorescence (Figure 15.90). These populations can easily be distinguished using a flow cytometer with both the 488 nm spectral line of an argon-ion laser and the 633 nm spectral line of a He–Ne laser for excitation. The kit components, number of assays and assay principles are summarized in Table 15.4. Apoptosis Assays Based on Protease Activity Caspases Caspases comprise a key component of the apoptotic machinery of cells, participating in an enzyme cascade that results in cellular disassembly. The recognition site for caspases is marked by three to four amino acids followed by an aspartic acid residue, with the cleavage occurring after the aspartate. These proteases are typically synthesized as inactive precursors. Inhibitor release or cofactor binding activates the caspase through cleavage at internal aspartates through autocatalysis or by the action of another protease. Caspase-3 Substrates and Assay Kits Caspase-3 is a key effector in the apoptosis pathway, amplifying the signal from initiator caspases (such as caspase-8) and signifying full commitment to cellular disassembly. In addition to cleaving other caspases in the enzyme cascade, caspase-3 has been shown to cleave poly(ADP-ribose) polymerase (PARP), DNAdependent protein kinase, protein kinase C and actin. Molecular Probes offers a selection of fluorogenic substrates (Table 15.5) containing the caspase-3 recognition site Asp-Glu-Val-Asp (DEVD); in particular our EnzChek Caspase-3 Assay Kits #1 and #2 provide a simple and direct assay of caspase-3 (Figure 15.91) and other DEVD-specific protease activities (e.g., caspase-7). Each kit contains: * Z-DEVD-AMC (in Kit E13183) or Z-DEVD-R110 (in Kit E13184) * Dimethylsulfoxide (DMSO) * Concentrated cell-lysis buffer * Concentrated reaction buffer * Dithiothreitol (DTT) * Ac-DEVD-CHO, a reversible aldehyde inhibitor * 7-Amino-4-methylcoumarin (AMC) (in Kit E13183) or rhodamine 110 (in Kit E13184) reference standard to quantitate the amount of fluorophore released in the reaction * Detailed protocols (EnzChek Caspase-3 Assay Kit #1 *Z-DEVD-AMC Substrate*, EnzChek Caspase-3 Assay Kit #2 *Z-DEVD-R110 Substrate*) Our EnzChek Caspase-3 Assay Kit #1 (E13183) contains the 7-amino-4-methylcoumarin (AMC)–derived substrate Z-DEVD-AMC () (where Z represents a benzyloxycarbonyl group). This substrate, which is weakly fluorescent in the UV spectral range (excitation/emission maxima ~330/390 nm), yields the blue–fluorescent product AMC (A191, Section 10.1, ), which has excitation/emission maxima of 342/441 nm upon proteolytic cleavage. The EnzChek Caspase-3 Assay Kit #2 (E13184) contains the rhodamine 110 (R110)–derived substrate, ZDEVD-R110 (). This substrate is a bisamide derivative of R110, containing DEVD peptides covalently linked to each of R110's amino groups, thereby suppressing both the dye's visible absorption and fluorescence. Upon enzymatic cleavage by caspase-3 (or a closely related protease), the nonfluorescent bisamide substrate is converted in a two-step process first to the fluorescent monoamide and then to the even more fluorescent R110 (R6479, Section 10.1, Figure 10.48, ). Both of these hydrolysis products exhibit spectral properties similar to those of fluorescein, with excitation/emission maxima of 496/520 nm. The Z-DEVD-R110 substrate (R22120) is also available separately in a 20 mg unit size for high-throughput screening applications. Either kit can be used to continuously measure the activity of caspase-3 and closely related proteases in cell extracts and purified enzyme preparations using a fluorescence microplate reader or fluorometer. The reversible aldehyde inhibitor Ac-DEVD-CHO can be used to confirm that the observed fluorescence signal in both induced and control cell populations is due to the activity of caspase-3–like proteases. Each of the kits contains sufficient reagents for about 500 assays using 100 µL volumes. Our EnzChek Caspase-3 Assay Kit #2 is also available as a convenient RediPlate 96 EnzChek Caspase-3 Assay Kit (R35100, RediPlate 96 EnzChek Caspase-3 Assay Kit), which includes one 96-well microplate, contained in a resealable foil packet to ensure the integrity of the fluorogenic components, plus all necessary buffers and reagents for performing the assay. The reversible aldehyde inhibitor Ac-DEVD-CHO is supplied in a separate vial for confirming the activity of caspase-3–like proteases. The enzyme sample to be assayed is added to the microplate in a suitable buffer, along with any compounds to be tested. Then, after incubation, the resultant fluorescence is quantitated on a fluorescence microplate reader equipped with filters appropriate for the green-fluorescent R110, with excitation/emission maxima of 496/520 nm. The microplate consists of twelve removable strips, each with eight wells, allowing researchers to perform only as many assay as required for the experiment (Figure 8.60). Eleven of the strips (88 wells) are preloaded with the Z-DEVDR110 substrate. The remaining strip, marked with blackened tabs, contains a dilution series of free R110 that may be used as a fluorescence reference standard. Table 10.3 summarizes our other RediPlate 96 and RediPlate 384 Assay Kits for protease activity (Section 10.4), phosphatase activity (Section 10.3) and dsDNA quantitation (Section 8.3). Significant discounts apply to purchases of multiple units of all of our RediPlate products. Also for assaying caspase-3 activity we offer Z-DEVD-AFC (A22121), which undergoes an ~65 nm red-shift to exhibit a peak emission of ~500 nm upon cleavage) and the bis-L-aspartic acid amide of R110 (D2-R110, R22122), which contains R110 flanked by aspartic acid residues (Table 15.5). D2-R110 does not appear to require any invasive techniques such as osmotic shock to gain entrance into the cytoplasm (). It may serve as a substrate for a variety of apoptosis-related proteases, including caspase-3 and caspase-7. Caspase-8 Substrates Caspase-8 plays a critical role in the early cascade of apoptosis, acting as an initiator of the caspase activation cascade. Activation of the enzyme itself is accomplished through direct interaction with the death domains of cell-surface receptors for apoptosis-inducing ligands. The activated protease has been shown to be involved in a pathway that mediates the release of cytochrome c from the mitochondria and is also known to activate downstream caspases, such as caspase-3. Three fluorogenic substrates containing the caspase-8 recognition sequence Ile-Glu-Thr-Asp (IETD) are available (Table 15.5); Z-IETD-AMC and Z-IETD-AFC (A22127, A22128; blue fluorescent after cleavage) and Z-IETD-R110 (R22125, R22126; green fluorescent after cleavage). Other Caspase and Granzyme B Substrates In addition to our R110-derived caspase-3 and -8 substrates, we offer R110-based substrates for caspase-1, -2, -6, -9 and -13, as well as substrates for granzyme B (Table 15.5). Granzyme B, a serine protease contained within cytotoxic T lymphocytes and natural killer cells, is thought to induce apoptosis in target cells by activating caspases and causing mitochondrial cytochrome c release. Cathepsins and Calpains The role of intracellular cathepsins and calpains in apoptosis is unclear, although an upstream role of cathepsin B in activation of some caspases and cathepsins during apoptosis has been established. Pepstatin A, which is a selective inhibitor of carboxyl (acid) proteases such as cathepsin D, has been reported to inhibit apoptosis in microglia, lymphoid cells and HeLa cells. Consequently, our cell-permeant BODIPY FL pepstatin derivative (P12271), which we have shown to inhibit cathepsin D in vitro (IC50 ~10 nM) and to target cathepsin D within lysosomes of live and fixed cells, may be of some utility in following the translocation of cathepsin D that may occur during apoptotic events. Calpains are a family of ubiquitous calcium-activated thiol proteases that are implicated in a variety of cellular functions including exocytosis, cell fusion, apoptosis and cell proliferation. Caspase-dependent downstream processing of calpain has been reported, suggesting that calpain may play a role in the degradation phase of apoptosis that is distinct from that of caspases. One mechanism of caspase dependence appears to be processing of the endogenous calpain inhibitor calpastin by caspase(s). However, calpain activation has also been reported to be upstream of caspases in radiation-induced apoptosis. Our t-BOC-Leu-Met-CMAC fluorogenic substrate (A6520) has been used to measure calpain activity in hepatocytes following the addition of extracellular ATP and may be of utility in detecting caspase-activated processing of procalpain in live single cells. Peptidase substrates based on our CMAC fluorophore (7-amino-4-chloromethylcoumarin, C2110; Section 10.1) passively diffuse into several types of cells, where the thiol-reactive chloromethyl group is enzymatically conjugated to glutathione by intracellular glutathione S-transferase or reacts with protein thiols, thus transforming the substrate into a membrane-impermeant probe. Subsequent peptidase cleavage results in a bright-blue–fluorescent glutathione conjugate (Section 10.4). Apoptosis Assays Using Mitochondrial Stains A distinctive feature of the early stages of programmed cell death is the disruption of active mitochondria. This mitochondria disruption includes changes in the membrane potential and alterations to the oxidation– reduction potential of the mitochondria. Changes in the membrane potential are presumed to be due to the opening of the mitochondrial permeability transition pore, allowing passage of ions and small molecules. The resulting equilibration of ions leads in turn to the decoupling of the respiratory chain and then the release of cytochrome c into the cytosol. Molecular Probes has available several unique reagents for studying changes in the mitochondria during apoptosis. * The green-fluorescent dye JC-1 (5,5',6,6'-tetrachloro-1,1',3,3'-tetraethylbenzimidazolylcarbocyanine iodide, T3168; ) exists as a monomer at low concentrations or at low membrane potential. However, at higher concentrations — aqueous solutions above 0.1 µM — or at higher membrane potentials, JC-1 forms redfluorescent "J-aggregates" (, , ) that exhibit a broad excitation spectrum and an emission maximum at ~590 nm (). Thus, the emission of this cyanine dye has been widely used to follow the changes in mitochondrial membrane potential that occur as a result of apoptosis. JC-1 has been used successfully to follow mitochondrial dysfunction in apoptotic hippocampal neurons and opening of the mitochondrial permeability transition pore (MTP). * Our JC-9 dye (3,3'-dimethyl--naphthoxacarbocyanine iodide, D22421, ) undergoes a similar potentialdependent spectral shift from a green-fluorescent product to a red-fluorescent aggregate () and is likely to be similarly useful for detecting apoptotic cells by both imaging and flow cytometry. Unlike JC-1, the green fluorescence of JC-9 is essentially invariant with membrane potential while the red fluorescence is significantly increased at hyperpolarized membrane potentials. * MitoTracker Red CMXRos (M7512, ) provides quick, easy and reliable detection of the loss of mitochondrial membrane potential that occurs during apoptosis. Our Patented MitoTracker Red CMXRos probe can be fixed using aldehyde-based fixatives and can thus be detected through subsequent immunocytochemistry, DNA end-labeling, in situ hybridization or counterstaining steps. The changes in mitochondrial membrane potential in osteosarcoma cells observed using MitoTracker Red CMXRos were instrumental in demonstrating the ability of these cells to undergo reversible apoptosis without entering cell death. Ratiometric measurements that compare the fluorescence of the membrane potential–dependent MitoTracker Red CMXRos label to that of the membrane potential–independent MitoTracker Green FM dye (M7514, Section 12.2) result in improved discrimination of apoptotic and non-apoptotic cell populations. * Rhodamine 123 (R302; FluoroPure Grade, R22420) is a cell-permeant, cationic, fluorescent dye that is readily sequestered by active mitochondria without inducing cytotoxic effects. Uptake and equilibration of rhodamine 123 is rapid — a few minutes — compared to other membrane potential–sensitive dyes, which may take 30 minutes or longer. Although not aldehyde-fixable, rhodamine 123 allows for quick and easy detection of apoptotic cells. * Most carbocyanine dyes with short (C1–C6) alkyl chains stain mitochondria of live cells when used at low concentrations (~0.5 µM or ~0.1 µg/mL). DiOC6(3) (D273) is a green-fluorescent cationic dye that accumulates in active mitochondria and is useful in following changes in the membrane potential of the mitochondria that occur during programmed cell death. This dye has been used in flow cytometric analysis to study mitochondrial changes in apoptotic human myeloid leukemia cells. * The accumulation of both the methyl and ethyl esters of tetramethylrhodamine (TMRM, T668; TMRE, T669) in mitochondria and endoplasmic reticulum is driven by membrane potential. TMRM has been used to study the temporal relationship between cytochrome c release from mitochondria and reduced mitochondrial membrane potential in apoptotic pheochromocytoma-6 cells and to investigate the mitochondrial permeability transition pore. * Calcein, a green-fluorescent dye that is formed inside cells that are loaded using calcein AM (C1430, C3099, C3100MP; Section 15.2, ), can be taken up into the matrix of mitochondria due to opening of the mitochondrial permeability transition pore (MTP). The MTP allows relatively large molecules (less than 620 daltons) to pass from the cytosol into the mitochondrial matrix. The transport of calcein through the MTP has been used to study the role of the MTP in apoptosis. * Nonyl acridine orange (A1372) is reported to bind to cardiolipin in the inner mitochondrial membrane. Its fluorescence decreases as cardiolipin becomes oxidized or otherwise altered during apoptosis. * Our SYTO 16 green-fluorescent nucleic acid stain (S7578) shows decreased fluorescence in apoptotic cells that may be due to changes in mitochondrial DNA conformation. It is optimally excited by the 488 nm spectral line of the argon-ion laser, making it useful for both flow cytometry and confocal laser-scanning microscopy. Apoptosis Assays Using Free Radical Probes The bcl-2 proto-oncogene product is reported to play a role in preventing apoptosis through its antioxidant properties. Following an apoptotic signal, cells sustain progressive lipid peroxidation — as detected with cisparinaric acid (P36005) — that can be suppressed by bcl-2 overexpression. cis-Parinaric acid was also used to assess lipid peroxidation in Down syndrome neurons, which exhibit increased levels of intracellular reactive oxygen species that lead to a reduction in levels of intracellular reduced glutathione and apoptosis. The reagent diphenyl-1-pyrenylphosphine (DPPP, D7894) is essentially nonfluorescent until it is oxidized by hydroperoxides to a phosphine oxide. Its lipid solubility makes DPPP similarly useful for detecting hydroperoxides in the membranes of live cells. Induction of apoptosis in human natural killer (NK) cells by monocytes is blocked by catalase, a scavenger of hydrogen peroxide, and by sodium azide, a myeloperoxidase inhibitor, whereas scavengers of superoxide and hydroxyl radicals do not prevent apoptosis. The most common fluorogenic probe for detecting reactive oxygen species is 2',7'-dichlorodihydrofluorescein diacetate (H2DCFDA, D399), which has been used to examine the effect of caspase-3 inhibitors on hydrogen peroxide production during apoptosis, in apoptotic embryos and in chemosensitive or chemoresistant cancer cells. H2DCFDA can detect so-called "necrotic zones" containing cells under oxidative stress in tissues; however, for this application we recommend our 5(and 6-)chloromethyl-2',7'-dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFDA, C6827; ), which has greater cell-membrane permeability and better cell retention of its green-fluorescent oxidation product. The acetoxymethyl ester of 2',7'-dichlorodihydrofluorescein diacetate (C2938, ) is also more permeant to live cells and tissues and has been used to detect hydrogen peroxide in transplanted myoblasts. As would be expected, the other major probes for reactive oxygen species — dihydrorhodamine 123 (D632, D23806; ) and dihydroethidium (hydroethidine; D1168, D11347, D23107), each of which is colorless and nonfluorescent until oxidized to the mitochondrial probe rhodamine 123 or to the nucleic acid stain ethidium — are also useful for detecting apoptotic cells in culture, and likely in tissues. Dihydrocalcein AM (D23805, ) is our newest scavenger of reactive oxygen species. The green-fluorescent dye calcein that is formed by intracellular oxidation () has cell retention that is superior to that of most other dyes (Figure 15.3). The principal oxidant of these probes is reportedly peroxynitrite, which is generated from nitric oxide (Section 19.3), although superoxide has also been implicated. Probes such as 10-acetyl-3,7-dihydroxyphenoxazine (the Amplex Red reagent, A12222, A22177; Section 19.2) react with hydrogen peroxide in the presence of a peroxidase to form red-fluorescent resorufin derivatives and may therefore be useful for correlating hydrogen peroxide production in cells with apoptosis. All of our probes for detecting reactive oxygen species are described in Chapter 19. Apoptosis Assays Using Ion Indicators Significant changes in intracellular pH, Na+, K+ and Ca2+ concentrations accompany apoptosis. The role of acidification in apoptosis has been investigated using carboxy SNARF-1 AM acetate (C1271, C1272; Section 21.2) and BCECF AM (B1150, B1170, B3051; Section 21.2) cell-permeant pH indicators. Low intracellular pH, as measured with the carboxy SNARF-1 pH indicator, and uptake of Hoechst 33342 have been shown to be correlated in partially apoptotic cell populations. Plasma membrane depolarization and inactivation of the Na+/K+-ATPase early in apoptosis leads to an increase in intracellular Na+ levels, as detected with SBFI AM (S1263, Section 22.1), and an inhibition of K+ uptake, as detected with 86Rb+ studies. Changes in intracellular Ca2+ levels may influence gene expression, as well as nuclease, protease and kinase activity. Our extensive selection of Ca2+ indicators, caged Ca2+ reagents, Ca2+ ionophores and Ca2+ chelators (Chapter 20) may help to sort out the mechanism of Ca2+ action in apoptosis. Apoptosis Assays Using Esterase Substrates Alterations in membrane permeability that occur during apoptosis have been monitored using nucleic acid stains (see above). These membrane changes may also affect the uptake and retention of our various general esterase substrates (Table 15.1) and substrates for other intracellular enzymes (Chapter 10). Results from staining apoptotic thymocytes with esterase substrates, however, showed significant variation depending on which probe was used. Some of this variation undoubtedly resulted from differences in the pH sensitivity of the probes; thus, calcein AM (C1430, C3099, C3100MP; Section 15.2), which has low pH sensitivity in the physiological pH range, may be the best reagent for detecting membrane permeability changes that accompany apoptosis. Calcein AM has been extensively used to detect the permeability transition of the mitochondrial membrane that apparently accompanies late stages of apoptosis. Calcein AM has been recommended as a better marker for early apoptotic events than annexin V conjugates in NIH 3T3 fibroblasts. Confocal laser-scanning microscopy of calcein AM–labeled cells shows a large increase in nuclear fluorescence and cell shrinkage during the early stages of chromatin condensation and nuclear fragmentation. Calcein AM staining also measures other important characteristics of apoptotic cells, including membrane blebbing and preservation of membrane integrity. An Apoptosis Assay that Measures the ATP:ADP Ratio Apoptotic cells are reported to have a relatively low ratio of ATP to ADP, apparently indicating decreased resynthesis of ATP in the mitochondria. A very sensitive chemiluminescent assay that measures the average ATP:ADP ratio of cultured apoptotic cells based on the principles of our luciferin/luciferase-based ATP Determination Kit (A22066; Section 10.3, Section 15.2) has been described.