Quantitative Variation
advertisement

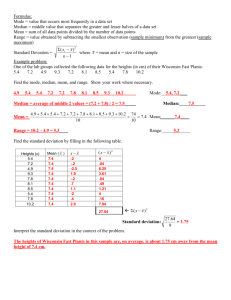
Biol 217 Evolution Spring 2010 Quantitative Variation in Helianthus annus (Sunflower) Seed Stripes Quantitative variation within and among groups is at the base of all studies of evolutionary change. In order to discuss changes in populations we need to develop a language and a set of concepts that allow us to describe and compare characteristics of populations. These skills involve understanding the distribution of characters in two broad categories: measures of central tendency; and, ways of describing the distribution of values around these measures of centrality. In this lab we will measure the extent of variation in a natural population within and among groups using the common sunflower (Helianthus annus) seed as the subject of our investigation. The language and concepts that we begin to explore today are the basis for making comparisons between different populations that exist at the same time and for examining changes in a population at different times. Stripe variation in the common sunflower (Helianthus annus) We will use histograms to represent the distribution of a population with respect to a particular character. In a histogram the character being discussed is represented on one axis while the number of individuals in the population with that value of the character are represented on the other axis. 1 Biol 217 Evolution Spring 2010 In this graph we see a representation of the following hypothetical data: #Stripes 0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 #Seeds 5 3 25 65 68 24 74 32 25 12 8 14 10 5 2 Part 1 – Collecting data Each student will receive three envelopes of BURPEE S Mammoth sunflower seeds. First, take a few minutes to spread them out and look at the ways they vary. In this first part of the activity you will divide up your sunflower seeds based on the number of stripes that they have. We will probably have some class discussion of how to score stripes. Keep them in separate piles based upon the number of stripes and photograph your results. Part 2 – Representing your data Record the number of seeds in each category (number of stripes). Then place them in test tubes arranged in a rack to create a histogram-like representation of your data. Compare your population to several other group’s populations. Then write 2-3 sentences describing your population with respect to the number of stripes. Part 3 – Measures of central tendency Calculate the mean, median and mode for your population. Show where each of these occurs by laying a piece of paper at the base of your test tubes and marking where each of these measures occur. Compare your results with a couple of other groups and write a sentence or 2 describing your population. mean = (∑ x)/n where x is a value and n is the number of values This statistic is also commonly referred to as the average. It is computed by adding each of the values in the distribution and dividing by the number of values in the distribution. 2 Biol 217 Evolution Spring 2010 Median: the middle value The median of a distribution can be found by putting the numbers in ascending order and taking the number in the middle, e.g. if the distribution has five values ( 5, 5, 4, 2, 1), then the third value is the median (4). If there is more than one middle value, then the mean of the two values is used. Mode: the most frequent value The mode is simply the value which occurs more often, e.g. the mode of this distribution, 8, 7 ,5 ,5, 5, 3, 1 is five. Mode Rat Weight Media n 7.5 5 Number of rats 10 Mean 2.5 1 2 3 4 6 5 7 8 9 10 Weight (g) Part 4 – Measures of distribution Enter your data into the JMP statistics program and have it calculate the standard deviation, skewness, and kurtosis values for your population distribution. Standard Deviation = √[∑ (x - "frequency mean")2/(n - 1) ] where x is a value and n is the number of values The standard deviation is a measure of the spread of a distribution. It is calculated by first calculating the mean of the distribution. Then, the difference between each value and the mean should be squared and all of those numbers added up. That value is divided by the total number of values minus one in the distribution and the square root of that number is the standard deviation. The standard deviation of a distribution indicates where most of the values are found. If 3 Biol 217 Evolution Spring 2010 you look at the values between the mean and one standard deviation above the mean 32% of the population values are represented (assuming that the population is normally distributed). We can extend this understanding of how the standard deviation describes a distribution to arrive at the following conclusions: 68% of the population falls within ±1 SD of the mean 95% of the population falls within ±2 SD of the mean 99.7% of the population falls within ±3 SD of the mean This is shown graphically on the figure below. Shell Length MEAN 40 20 NUMBER 30 10 10 20 30 40 Shell Length ± 1- 68% ± 2- 95% ± 3- 99.7% Skewness = (1/ns3)∑(x - "frequency mean")3 where s is the std. deviation, x and n as before Skewness is a statistic that describes the relative sizes of the tails of the distribution. A negative skewness value implies that the left tail of the distribution is longer. A positive skewness value implies that the right tail of the distribution is longer. The calculation is similar to calculations for the standard deviation. After finding the frequency mean, the cubes of the differences between the each of the values and the mean are summed. Then, that number is divided by the number of values in the distribution and the cube of the standard deviation. 4 Biol 217 Evolution Spring 2010 Fruit Weight 30 NUMBER 20 10 1 2 3 4 5 6 7 8 9 10 Fruit Weight A positively skewed distribution. The red line represents a symmetrical distribution for reference. Note that in a positively skewed distribution the mean will be greater than the median population value. Kurtosis = (1/ns4)∑(x - "frequency mean")4 - 3 s, n and x as before Kurtosis is a statistic that describes how sharp the peak of the distribution is. A negative score indicates platykurtosis (a relatively flat peak), while a positive score indicates leptokurtosis (a relatively sharp peak). mesokurtic "normal" platykurtic 300 100 leptokurtic 10 20 30 X 5 Number 200 Biol 217 Evolution Spring 2010 Part 5 – Multidimensional variation Using the seeds from within one category of stripe number, make three additional sets of measurements on your seeds. For example, if you looked at all 19 seeds that had 11 stripes (hypothetical data), you also could measure seed weight, buoyancy, seed length, oil extraction, etc. to examine intraclass variation. Lab Write up (Individual) In addition to submitting your JMP file electronically, your write up should include: Your definition for the character stripe number. Include two photos of an individual seed (i.e., one picture of each flat side) and clearly mark the stripes that you counted in your analysis. Your raw data for variation with respect to stripe number in your population in tabular form. Include a photograph of the histogram made by arranging your seeds in test tubes of equal size. A clearly labeled graph of the distribution of your population on which you have indicated the mean, median, mode (with reference to the histogram) and the calculated values for standard deviation, skew and kurtosis. A clearly labeled graph of the distribution of the full class’s population. A short description of your data (not simply listing the statistics but describing it as you might to a friend who was not familiar with statistics) and comparison of your data to another group’s data. Do you think they are both samples from the same population? Why or why not? Discuss why central tendency may not be such an important factor for natural historian with a Darwinian perspective. A definition of the three other characters that you measured. A description of your findings on interclass variation including a histogram of your results. Show a scatter plot with a linear regression line for the most correlated of two of your three quantitative traits that you measured on seeds with the same number of stripes. A brief discussion of how this detailed information about variation is relevant to evolution. Sunflowers are an important economic crop. How has natural and artificial selection contributed to the variation that you have observed? Several references and one pedigree are presented below that may help you to think about this problem. 6 Biol 217 Evolution Spring 2010 This is a modified version of a lab developed by John Jungck at Beloit College. Several of the figures were taken from the Biometrics module by Daniel Hornbach published the BioQUEST Library. See http://bioquest.org/biostat for additional information about these statistics. A more detailed explanation of how to calculate these statistics can be found in Biometry by Sokal and Rohlf, which is the text for the Beloit College Biometrics course. Chapter 4, Desriptive Statistics, gives a good explanation of the first four statistics. Chapter 2 is on frequency distributions and may be of some help. Tang et al. (2006) reported the above data and three loci model for the inheritance of sunflower seed characteristics. Considerable variation exists in seed characteristics of different cultivars of sunflower. A sample of this variation from a seed company in China is illustrated below. 7 Biol 217 Evolution Spring 2010 Newfield.com source: Ningxia Newfield Foods Co., Ltd., China http://www.newfield.com.cn/english/products/sunflowerseeds.htm References: Bert, P.F., I. Jouan, D. Tourvielle de Labrouche, F. Serre, J. Philippon, P. Nicolas, and F. Vear. (2003). Comparative genetic analysis of quantitative traits in sunflower (Helianthus annuus L.). 2. Characterization of QTL involved in developmental and agronomic traits. Theoretical and Applied Genetics 107:181–189. Smith, J. Stephen C., Eric Hoeft, Glenn Cole, Henry Lu, Elizabeth S. Jones, Steven J. Wall, and Donald A. Berry. (2009). Genetic Diversity among U.S. Sunflower Inbreds and Hybrids: Assessing Probability of Ancestry and Potential for Use in Plant Variety Protection. Crop Science 49: 1295-1303. Tang, Shunxue, Alberto Leon, William C. Bridges, and Steven J. Knapp. (2006). Quantitative Trait Loci for Genetically Correlated Seed Traits are Tightly Linked to Branching and Pericarp Pigment Loci in Sunflower. Crop Science 46: 721-734. 8 Biol 217 Evolution Spring 2010 Other related references of possible use for your analysis and report: Angadi, S. V., and M. H. Entz. (2002). Water Relations of Standard Height and Dwarf Sunflower Cultivars. Crop Science 42: 152-159. Lambrides, C. J., S. C. Chapman, and R. Shorter. (2004). Genetic Variation for Carbon Isotope Discrimination in Sunflower: Association with Transpiration Efficiency and Evidence for Cytoplasmic Inheritance. Crop Science 44: 16421653. Lenardon, S. L., M. E. Bazzalo, G. Abratti, C. J. Cimmino, M. T. Galella, M. Grondona, F. Giolitti, and A. J. León. (2005). Screening Sunflower for Resistance to Sunflower chlorotic mottle virus and Mapping the Rcmo-1 Resistance Gene. Crop Science 45: 735-739. Robinson, R. G., L. A. Bernat, H. A. Geise, F. K. Johnson, M. L. Kinman, E. L. Mader, R. M. Oswalt, E. D. Putt, C. M. Swallers, and J. H. Williams. (1967). Sunflower Development at Latitudes Ranging from 31 to 49 Degrees. Crop Science 7: 134-136. Ruiz, Ricardo Adolfo, and Gustavo Angel Maddonni. (2006). Sunflower Seed Weight and Oil Concentration under Different Post-Flowering Source-Sink Ratios. Crop Science 46: 671-680. Tang, Shunxue, Adam Heesacker, Venkata K. Kishore, Alberto Fernandez, El Sayed Sadik, Glenn Cole, and Steven J. Knapp. (2003). Genetic Mapping of the Or5 Gene for Resistance to Orobanche Race E in Sunflower. Crop Science 43: 1021-1028. de la Vega, Abelardo J., and Scott C. Chapman. (2006). Defining Sunflower Selection Strategies for a Highly Heterogeneous Target Population of Environments. Crop Science 46: 136-144. Yu, Ju-Kyung, Shunxue Tang, Mary B. Slabaugh, Adam Heesacker, Glenn Cole, Martin Herring, John Soper, Feng Han, Wen-Chy Chu, David M. Webb, Lucy Thompson, Keith J. Edwards, Simon Berry, Alberto J. Leon, Martin Grondona, Christine Olungu, Nele Maes, and Steven J. Knapp. (2003). Towards a Saturated Molecular Genetic Linkage Map for Cultivated Sunflower. Crop Science 43: 367-387. Yue, Bing, Xiwen Cai, Brady A. Vick, and Jinguo Hu. (2009). Genetic Diversity and Relationships among 177 Public Sunflower Inbred Lines Assessed by TRAP Markers. Crop Science 49: 1242-1249. 9 Biol 217 Evolution Spring 2010 10 Biol 217 Evolution Spring 2010 Notes to Teaching Assistants: [Intro to JMP: columns, variables, distributions, changing axes, looking at moments. Have them enter their data into JMP and generate a graph of the distribution of their population. Print it. [print preview} Put their print out on paper in front of test tubes Rough time estimates: 10-15 minutes intro to measures of distribution 10-15 minute intro to JMP Half hour collecting data Half hour: They enter data, do calculations, print graphs. Class discussion: [put up some data on board as well as some of the means and see if that is a useful way to describe populations for comparisons.] Range Standard deviation mean ±1 SD is 68% of population; ±2SD is 95% of population; ±3SD is 99.7% of pop. Skewedness +skew is shifted left or pulled right (mean is greater than the median) –skew is shifted right or pulled left (mean is less that the median) Kurtosis – is platykurtic (flat); + is leptokurtic (pointy); and 0 is mesokurtic (normal). 11