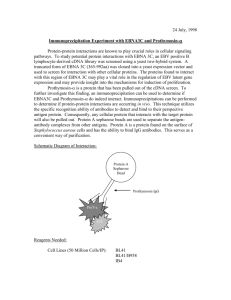
RNA::Protein Immunoprecipitation RNA Pulldown
advertisement
Molecular Techniques and Methods RNA::Protein Immunoprecipitation RNA Pulldown Copy Right © 2001/ Institute of Molecular Development LLC INTRODUCTION Formaldehyde crosslinks RNA::Protein and protein::protein interactions. After crosslink, RNA::protein complex is immunoprecipitated and RNA is released. All works should be done in RNase-free condition. MATERIALS AND SOLUTIONS EMSA Buffer (50 ml) 25mM HEPES ---------------------------------------- 1.25 ml of 1 M 3mM MgCl2 ----------------------------------------- 150 ul of 1 M 150mM KCl ----------------------------------------- 7.5 ml of 1 M 1mM DTT ------------------------------------------- 50 ul of 1 M 5mg/ml Heparin ------------------------------------ 250 mg 5% glycerol --------------------------------------- 5 ml of 50% 0.05% Tween 20 ------------------------------------ 25 ul of 100% DW to make a final volume of ---------------------- 50 ml 10x DNase Buffer (1 ml) 250mM MgCl2 ---------------------------------------- 250 ul of 1 M 50mM CaCl2 ----------------------------------------- 50 ul of 1 M DEPC water ----------------------------------------- 700 ul 40 mM Vanadyl ribonucleoside complexes (1 ml) Vanadyl ribonucleoside complexes ------------------- 20 mg DEPC-H2O ----------------------------------------- 1 ml Protein-RNA Elution Buffer (10 ml) 100mM Tris-HClpH 8.0 ----------------------------- 1 ml of 1 M 10mM Na2-EDTA -------------------------------------- 200 ul of 0.5 M 1% SDS --------------------------------------------- 1 ml of 10% DEPC-water ----------------------------------------- 8 ml PROCEDURES Bead Preparation a. Using a large orifice pipet tip, place 50-100 ul Protein A/G beads slurry into four 1.5ml centrifuge tubes. (50% A/G slurry in PBS preabsorbed by BSA) b. Add 450 ul EMSA Buffer to each tube. Flick to mix and centrifuge for 30 seconds at 10,000xg. c. Use a gel loading tip to aspirate all of the supernatant. Wash once more with 500 ul EMSA Buffer. d. After removing final wash, resuspend the beads in 50 ul EMSA Buffer. e. To two tubes, add 1 ~ 5 ug of antibody of interest. The amount of antibody can be decreased to lower background. To the other two tubes add 1 ~ 5 ug IgG specific to the species used in the antibody. For example: If primary antibody is mouse monoclonal, the IgG used must be mouse IgG for comparison. f. Incubate beads and antibody at 4°C on the rocker for at least 4 hours. Incubation time can be increased or decreased for different results. Every so often, flick each tube to make sure that the slurry is well mixed. In cell crosslink between Protein::RNA complexes 1. Ten petridishes (10 cm diameter) of cell culture generate enough cells for IP/RT-PCR. Alternatively, 6 of T175 plates with 90% cell confluency is necessary. Cell pellet volume needs to be about 0.5 ml after collection. 2. Treat trypsin for the shortest time as possible (3-5 min). 3. Add double volume (Medium + 10% serum) to end the digestion, collect cells, spin at 400g for 5 min at RT. 4. Wash the cell pellet with 10 ml cold PBS. Spin at 400g for 5 min at 4oC. 5. Repeat step 4. 6. Resuspend cell pellet in 10 ml PBS. 7. Add 270 ul of 37% formaldehyde. 8. Incubate at RT or 37oC for 15-20 min on rocking platform. Cells may be incubated longer at 4oC. Alternatively, cells are irradiated under UV light for RNA::protein cross-linking. 9. To stop cross-linking, add 1.4 ml sterile 2 M glycine (final conc = 0.2 M) and incubate 5 min at RT or 37oC on rocking platform. 10. Spin at 400g for 5 min at 4oC. 11. Wash with 10 ml cold PBS. 2-3x. Cell Lysate Preparation 12. For one experiment usually 2 previously formaldehyde cross-linked cell pellets is enough. This equates to about 5-6 T175 flasks of cells. 13. Follow the Pierce NE-PER Nuclear and Cytoplasmic protein extraction kit directions, multiplying all reagents by 5: 14. Combine two tubes of pellets with 1mL of CER I + Protease Inhibitor. 15. Mix well and incubate on ice for 10 minutes. 16. Add 55 µL CER II. Vortex, incubate on ice 1 minute, vortex again and centrifuge for 5 minutes at 4°C at 16,000 x g. 17. Remove supernatant (cyto fraction) and store on ice. Resuspend pellet in 500 µL NER + Protease Inhibitor. 18. Vortex every 10 minutes for a total of 40 minutes. Incubate on ice in between vortexing. 19. Centrifuge at 16,000g for 10 minutes at 4°C, save supernatant (nuc fraction) on ice. Discard pellet. 20. Sonicate each lysate at 50% amplitude. Sonicate for 15 seconds on ice, rest for 2 minutes on ice, then sonicate another 15 seconds on ice. 21. Remove 500 µL cyto fraction and store at -80oC for future use. 22. To the 500 µL of Cyto and Nuc cell lysates add: 10x DNase Buffer 50 ul RNase Inhibitor 40 mM Vanadyl ribonucleoside complexes RNase-free DNase 2.5 ul 5 ul 5 ul For RNase treatment, do at this step. 23. Incubate at 37°C for 15 minutes. 24. Add 20µL 0.5M EDTA to stop the reaction. 25. Centrifuge 5 minutes at 14,000 rpm at 4oC. Collect supernatant and store on ice or at -70oC. Bead/Lysate Incubation 26. After beads have been incubate with antibody, wash 5 times with EMSA Buffer. 27. First spin down beads at 10,000g for 30 seconds. Aspirate supernatant using a very fine tipped gel loading tip. 28. Wash with 500 µL EMSA Buffer. Repeat for a total of 5 washes. 29. After the last wash, add 50 µL EMSA Buffer to the beads. Add 250 µL of each cell lysate to the beads so you will have the following samples: Antibody/Cyto IgG/Cyto Antibody/Nuc IgG/Nuc 30. Incubate overnight at 4oC on the rocker. Protein/RNA Elution 31. Wash beads 5 times with EMSA Buffer as previously described. 32. After last wash, remove all supernatant and add 100 µL protein-RNA elution buffer. 33. Incubate at 37oC for 10 minutes. 34. Centrifuge at room temperature for 2 minutes at 3000rpm. 35. Save supernatant (eluant). 36. Repeat the elution with another 100µL protein-RNA elution buffer and pool elutants. 37. Save 150 µL eluant on ice for RNA extraction. Use remaining eluant (usually 20-35 µL) to run the SDS-PAGE and Western. 38. Divide amount of eluant used (use the same for each sample) by 4 to determine the amount of 5x Sample Buffer to use. 39. Add sample buffer to eluants, incubate 5 minutes at 95oC, and load into gel along with a positive control. 40. Continue with Western protocol. RNA Purification 41. To 150 µL eluants, add 6µL 5M NaCl and 20 µL Proteinase K. 42. Incubate for 30 minutes at 55oC. Flick every 10 minutes to mix. 43. Add 100 µL DEPC water. 44. To each, add 245 µL phenol:CHCl3 and 5 µL IAA (3-methyl 1-butanol, anhydrous). 45. Mix and centrifuge 5 minutes at 13,000 rpm at 4oC. Remove the upper layer and save. Discard lower layer. 46. To the upper layer, add 240 µL CHCl3 and 10 µL IAA. 47. Mix and centrifuge 5 minutes at 13,000 rpm at 4oC. Remove the upper layer and save. Discard lower layer. 48. To the upper layer, add 25µL 3M sodium acetate (RNA shelf, RT), 4µL glycogen , and 650µL 100% ethanol (flam cab) 49. Precipitate RNA at -20oC overnight, or longer if necessary. 50. Centrifuge RNA for 50 minutes at 4oC at 13,000 rpm. 51. Make sure that there is a pellet in each tube. If not, continue precipitation at -20oC. Otherwise, pour off supernatant being careful not to disturb the cell pellet. 52. Centrifuge another 10 minutes at 4oC at 13,000 rpm. Carefully remove supernatant from RNA pellet. cDNA Synthesis and qRT-PCR 53. Do as in the qRT-PCR.. 10 ul cDNA synthesis reaction is enough. Dilute cDNA reation by adding 100 ul DEPC-H2O for qPCR reaction. NOTES Things that can be changed/optimized: 1. Incubation time of beads with antibody a. 2 hours to overnight at 4C b. 30 minutes at RT 2. Amount of antibody added to beads a. Decrease in antibody gives decrease in background 3. RNA precipitation a. Overnight or 2 days at -20C b. Could possibly use Trizol protocol 4. Amount of lysate added to beads a. Between 150µl-500µl lysate used. Less will lower background, more will increase sensitivity. KIT INFORMATION REFERENCES Niranjanakumari S. et al. (2002) Reversible cross-linking combined witrh immunoprecipitation to study RNA-preotein interactions in vivo. Methods 26, 182-190. Keene JD et al. (2006) RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes fromo cell extracts. Nature Protocols. 1. 302-307. Tiina Peritz et al. (2006) Immunoprecipitation of mRNA-protein complexes. Nature Protocols. 1, 577-580. Somashe Niranjanakumari et al. (2002) Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 26, 182-190. Please send your comment on this protocol to "editor@MolecularInfo.com". Home MT&M Online Journal Hot Articles Order Products Classified