Wellcome Trust Combined Training Programme 12 week Projects
advertisement
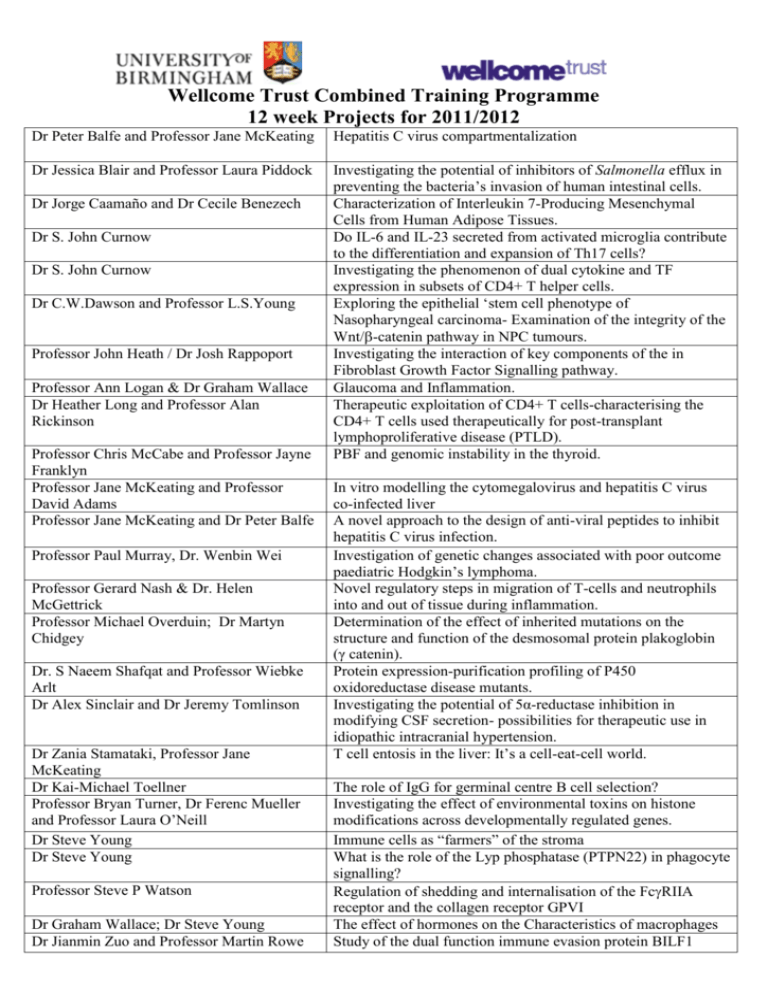
Wellcome Trust Combined Training Programme
12 week Projects for 2011/2012
Dr Peter Balfe and Professor Jane McKeating
Hepatitis C virus compartmentalization
Dr Jessica Blair and Professor Laura Piddock
Investigating the potential of inhibitors of Salmonella efflux in
preventing the bacteria’s invasion of human intestinal cells.
Characterization of Interleukin 7-Producing Mesenchymal
Cells from Human Adipose Tissues.
Do IL-6 and IL-23 secreted from activated microglia contribute
to the differentiation and expansion of Th17 cells?
Investigating the phenomenon of dual cytokine and TF
expression in subsets of CD4+ T helper cells.
Exploring the epithelial ‘stem cell phenotype of
Nasopharyngeal carcinoma- Examination of the integrity of the
Wnt/-catenin pathway in NPC tumours.
Investigating the interaction of key components of the in
Fibroblast Growth Factor Signalling pathway.
Glaucoma and Inflammation.
Therapeutic exploitation of CD4+ T cells-characterising the
CD4+ T cells used therapeutically for post-transplant
lymphoproliferative disease (PTLD).
PBF and genomic instability in the thyroid.
Dr Jorge Caamaño and Dr Cecile Benezech
Dr S. John Curnow
Dr S. John Curnow
Dr C.W.Dawson and Professor L.S.Young
Professor John Heath / Dr Josh Rappoport
Professor Ann Logan & Dr Graham Wallace
Dr Heather Long and Professor Alan
Rickinson
Professor Chris McCabe and Professor Jayne
Franklyn
Professor Jane McKeating and Professor
David Adams
Professor Jane McKeating and Dr Peter Balfe
Professor Paul Murray, Dr. Wenbin Wei
Professor Gerard Nash & Dr. Helen
McGettrick
Professor Michael Overduin; Dr Martyn
Chidgey
Dr. S Naeem Shafqat and Professor Wiebke
Arlt
Dr Alex Sinclair and Dr Jeremy Tomlinson
Dr Zania Stamataki, Professor Jane
McKeating
Dr Kai-Michael Toellner
Professor Bryan Turner, Dr Ferenc Mueller
and Professor Laura O’Neill
Dr Steve Young
Dr Steve Young
Professor Steve P Watson
Dr Graham Wallace; Dr Steve Young
Dr Jianmin Zuo and Professor Martin Rowe
In vitro modelling the cytomegalovirus and hepatitis C virus
co-infected liver
A novel approach to the design of anti-viral peptides to inhibit
hepatitis C virus infection.
Investigation of genetic changes associated with poor outcome
paediatric Hodgkin’s lymphoma.
Novel regulatory steps in migration of T-cells and neutrophils
into and out of tissue during inflammation.
Determination of the effect of inherited mutations on the
structure and function of the desmosomal protein plakoglobin
(γ catenin).
Protein expression-purification profiling of P450
oxidoreductase disease mutants.
Investigating the potential of 5α-reductase inhibition in
modifying CSF secretion- possibilities for therapeutic use in
idiopathic intracranial hypertension.
T cell entosis in the liver: It’s a cell-eat-cell world.
The role of IgG for germinal centre B cell selection?
Investigating the effect of environmental toxins on histone
modifications across developmentally regulated genes.
Immune cells as “farmers” of the stroma
What is the role of the Lyp phosphatase (PTPN22) in phagocyte
signalling?
Regulation of shedding and internalisation of the FcRIIA
receptor and the collagen receptor GPVI
The effect of hormones on the Characteristics of macrophages
Study of the dual function immune evasion protein BILF1
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Dr Peter Balfe and Professor Jane McKeating
Contact details: p.balfe@bham.ac.uk
Office Location: Institute for Biomedical Research office 535
School: Immunity and Infection
Hepatitis C virus compartmentalization
Outline of Project:
Hepatitis C Virus (HCV) poses a global health problem, with over 170 million infected individuals
worldwide at risk of developing cirrhosis and hepatocellular carcinoma. Current therapies are limited and
recent trials of direct acting anti-viral agents select for drug resistant strains, demonstrating the need for
combination therapies of drugs targeting different steps of the viral replication cycle. Recent observations
that a minority (10-15%) of hepatocytes express viral antigens in the liver1 and HCV can transmit via cellcell contacts2,3, support a model where HCV replicates at distinct sites in the liver. Cell-to-cell spread limits
HCV trafficking and the resulting viruses follow independent evolutionary histories, i.e. become compartmentalized. Genetic
models predict that evolution will occur more rapidly in small isolated populations than in a homogeneous
pool. We hypothesize that some areas of the liver are inherently more able to control HCV
replication than others. This project will profile HCV and cellular mRNA quantities encoding key
inflammatory mediators in multiple biopsies collected from infected liver. These studies will identify key
inflammatory pathways that control HCV replication in vivo and will inform the design of future therapies.
Techniques to be used in the project: The student will be trained in the preparation of cellular RNA and
quantitative real time PCR array technologies to measure the abundance of mRNA species encoding a
defined number of inflammatory mediators. These techniques will provide a generic skill base that is widely
applicable to a range of biologically diverse projects.
Are these techniques already established in the laboratory where the project would take place? Yes
Names of other researchers who may also be involved in project (whether supervisory or technical):
Drs Luke Meredith and Ian Rowe, hepatitis C virologists.
Key References:
1. Liang Y, et al. 2009. Gastroenterology 137: 1448-1458.
2. Timpe JM, et al. 2008. Hepatology 47: 17-24.
3. Brimacombe CL, et al. 2011. Journal of Virology 85: 596-605.
Does this project require the student to hold a personal home office licence? No
Do you have all the relevant ethical approval for this study? Yes
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Dr Jessica Blair and Professor Laura Piddock
Contact details: J.M.A.Blair@bham.ac.uk
Office Location: IBR West Extension
School: School of Immunity and Infection
Investigating the potential of inhibitors of Salmonella efflux in preventing the bacteria’s invasion of
human intestinal cells.
Outline of Project:
Salmonella enterica is a major cause of gastrointestinal infection and systemic bacteraemia in humans
worldwide and is the cause of 1.3 million deaths each year. Multi-drug resistance (MDR) in this important
human pathogen is an increasing problem. Bacteria, such as Salmonella, have efflux pump systems that
pump antibiotic compounds out of bacterial cells and over-expression of some of these efflux pumps such as
AcrAB-TolC can confer multi-drug resistance to clinically useful antibiotics. In addition efflux pumps,
including AcrAB-TolC, are required for the ability of various pathogens to cause infection. Genetic
inactivation of components of the AcrAB-TolC system makes bacteria more susceptible to antibiotics and
also less virulent. The role of RND efflux systems in both antimicrobial resistance and virulence makes
them attractive targets for the design of efflux pump inhibitors (EPIs) which could be used clinically, in
combination with antimicrobials to enhance and preserve clinical effectiveness and potentially also to act as
anti-virulence compounds. Work in our laboratory has shown that one EPI, PAβN, inhibited virulence of
Salmonella and the aim of this project is to determine whether all inhibitors of efflux prevent invasion of
human intestinal cells by Salmonella and could therefore act as anti-virulence drugs. This information is
vital for design of future EPIs, possibly active against multiple bacterial pathogens, and the development of
successful antibiotic and EPI regimens.
Techniques to be used in the project: This project will involve training in the following:
-
Basic microbiology
Tissue culture of human intestinal cells (including infection assays with Salmonella)
Molecular biology techniques (including PCR, sequencing etc)
Are these techniques already established in the laboratory where the project would take place?
Yes
Names of other researchers who may also be involved in project (whether supervisory or technical):
Key References:
1. Piddock, L.J.V. (2006). Bacterial Multi-Drug Efflux Pumps – not just for resistance. Nature Microbiology
Reviews. 4: 629-636
2. Blair, J.M.A., La Ragione, R.M., Woodward, M.J. and Piddock, L.J.V. (2009). Periplasmic adaptor
protein AcrA has a distinct role in the antibiotic resistance and virulence of Salmonella enterica serovar
Typhimurium JAC. 64(5):965-72.
3. Buckley, A. M., M. A. Webber, S. Cooles, L. P. Randall, R. M. La Ragione, M. J. Woodward and L. J. V.
Piddock (2006). The AcrAB-TolC System plays a role in pathogenesis. Cellular Micro 8(5): 847-856.
Does this project require the student to hold a personal home office licence?
No
Do you have all the relevant ethical approval for this study?
NA
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Dr Jorge Caamaño and Dr Cecile Benezech
Contact details: j.caamano@bham.ac.uk and c.benezech@bham.ac.uk
Office Location: IBR bldg. Room 437
School: Immunity and Infection
Characterization of Interleukin 7-Producing Mesenchymal Cells from Human Adipose Tissues
Outline of Project:
One of the characteristics of obesity is the production of inflammatory cytokines. Adipose tissues
contain T cells and macrophages that contribute to the general inflammatory status. Our recent work on
mouse adipose tissues has identified a population of stromal cells that produces Interleukin-7 and other
factors that support T cell survival ex vivo1-3. During this project the student will isolate stromal cells from
human tissues and characterize the expression of specific cellular markers involved in the recruitment of T
cells and macrophages. Moreover, the mechanism by which TNF-a stimulation induces the expression of T
cell survival factors on these cells will be investigated. Our working hypothesis is that stromal cells in
adipose tissues expressing IL-7, chemokines, and cell adhesion molecules are essential for the
recruitment and survival of immune cells in adipose tissues to induce inflammation during obesity.
The student will work in close collaboration with a postdoctoral researcher and PhD students in our group.
She/he will receive training in cell and molecular biology techniques, induction and quantification of
adipocyte differentiation, RNA preparation, and to assess gene expression by QPCR. The student will be
able to discuss his/her data during informal daily meetings, and more formally in weekly meetings.
Techniques to be used in the project: Cell culture, FACs analysis, high speed cell sorting, PCR and
QPCR, fluorescence and confocal microscopy.
Are these techniques already established in the laboratory where the project would take place?
Yes
Names of other researchers who may also be involved in project (whether supervisory or technical):
Anita Challa, Razvan Dolea, Omar Qureshi, Prof. Dave Samson.
Key References:
1- C. Bénézech, E. Mader, M. Khan, K Nakamura, A. White, C. Ware, G. Anderson and J. Caamaño. (2011). Adipocyte Precursor
Cells Give Rise to Lymph Node Stromal Cells. Immunity (In Revision).
2- Bénézech C, White A, Mader E, Serre K, Parnell S, Pfeffer K, Ware CF, Anderson G, Caamaño JH. (2010). Ontogeny of
stromal organizer cells during lymph node development. J Immunol. 184:4521-30.
3- White, A., et al. (2007) Lymphotoxin a-dependent and -independent signals regulate stromal organizer cell homeostasis during
lymph node organogenesis. Blood 110, 1950-1959
Does this project require the student to hold a personal home office licence?
No
Do you have all the relevant ethical approval for this study?
Yes
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Dr S. John Curnow
Contact details: s.j.curnow@bham.ac.uk
Office Location: University Research Labs, QE Hospital
School: I&I
Do IL-6 and IL-23 secreted from activated microglia contribute to the differentiation and expansion
of Th17 cells?
Outline of Project:
Background. Multiple sclerosis (MS) is a chronic inflammatory condition with a significant
autoimmune component. Data from both animal models and patients with MS suggest that CD4+ Th17 cells
are involved in disease pathogenesis. The differentiation of Th17 cells is dependent on a number of
cytokines including IL-6 and IL-23. The cellular source of these cytokines in MS is not known, but may
involve microglia (a type of macrophage) within the central nervous system. Such a finding would offer
microglia as suitable cellular targets to pharmacologically manipulate for the therapeutic benefit of patients
with MS.
Research question. We hypothesis that IL-6 and IL-23 secreted from activated microglia will contribute to
the differentiation and expansion of Th17 cells.
Study design and methods. The expression of IL-6 and IL-23 from a human microglial cell line, following
activation, will be determined by qRT-PCR and ELISA. CD4+ T cells will be purified from healthy human
peripheral blood and co-cultured with the activated microglia. The frequency of Th17 cells will be
determined by measuring intracellular cytokine levels following stimulation.
Techniques to be used in the project:
Primary cell purification, cell culture, multi-colour flow cytometry, qRT-PCR, ELISA.
Are these techniques already established in the laboratory where the project would take place?
Yes/No
Names of other researchers who may also be involved in project (whether supervisory or technical):
Prof. Nicholas M. Barnes, Siobhan Restorick, Lindsay Durant
Key References:
1. Gandhi R et al. Role of the innate immune system in the pathogenesis of multiple sclerosis.
J Neuroimmunol. 2010 221:7-14.
2. Korn, T. et al. IL-17 and Th17 Cells. Annu. Rev. Immunol. 2009 27: 485-517.
3. Li Y et al. Inducible IL-23p19 expression in human microglia via p38 MAPK and NF-kappaB signal
pathways. Exp Mol Pathol. 2008 84:1-8.
Does this project require the student to hold a personal home office licence?
Yes/No
Do you have all the relevant ethical approval for this study?
Yes/No
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Dr S. John Curnow
Contact details: s.j.curnow@bham.ac.uk
Office Location: University Research Labs, QE Hospital
School: I&I
Investigating the phenomenon of dual cytokine and TF expression in subsets of CD4+ T helper cells.
Outline of Project:
Background. CD4+ T cells differentiate into a number of distinct T helper (Th) lineage, each defined by the
production of specific cytokines and expression of lineage-defining transcription factors (TF). Examples
include Th1 cells expressing T-bet and secreting IFN, and Th17 cells that secrete IL-17 and express
RORC. However, there are some Th cells that secrete more than one lineage-defining cytokine (e.g. IFN in
combination with IL-17). We have recently shown that these dual cytokine-secreting cells express both
lineage-defining transcription factors.
Research question. We wish to determine whether the phenomenon of dual cytokine and TF expression is
universal, by studying the combination of other Th subsets, specifically Th1 with Th2 (IL-4) and Th1 with
Tr1 (a regulatory cell secreting IL-10).
Study design and methods. CD4+ T cells will be purified from healthy human peripheral blood and cultured
under Th lineage polarising conditions, biased towards either Th2 or Tr1 cells. The expression of lineagedefining cytokines and TF will be measured using multi-colour flow cytometry and quantitative RT-PCR
analysis.
Techniques to be used in the project:
Primary cell purification, cell culture, multi-colour flow cytometry and qRT-PCR.
Are these techniques already established in the laboratory where the project would take place?
Yes/No
Names of other researchers who may also be involved in project (whether supervisory or technical):
Siobhan Restorick, Lindsay Durant
Key References:
1. Mazza G et al. Isolation and characterization of human interleukin-10-secreting T cells from peripheral
blood. Hum Immunol. 2010 71:225-34.
2. Korn, T. et al. IL-17 and Th17 Cells. Annu. Rev. Immunol. 2009 27: 485-517.
Does this project require the student to hold a personal home office licence?
Yes/No
Do you have all the relevant ethical approval for this study?
Yes/No
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Dr C.W.Dawson and Professor L.S.Young
Contact details: Tel: 414-4484 e-mail: c.w.dawson@bham.ac.uk
Office Location: Room Ground floor clinical research block
School: Cancer Sciences
Exploring the epithelial ‘stem cell phenotype of Nasopharyngeal carcinoma- Examination of the integrity of the
Wnt/-catenin pathway in NPC tumours.
Outline of Project:
Unlike other squamous carcinomas of the head and neck (HNSCC), the poorly differentiated and
undifferentiated forms of NPC are consistently associated with EBV infection (1). This has led to
speculation that NPC may originate from populations of undifferentiated early progenitor cells such as stem
cells or cancer stem cells (CSCs)(2). Gene expression profiling of NPC tumours has identified dysregulated
expression of the Sonic hedgehog (Shh), Wnt/-catenin and Transforming growth factor- (TGF/Bone
morphogenic protein (BMP) signalling pathways, findings which are intriguing given that these pathways
are implicated in stem cell/CSC maintenance (2). The aim of this project is to examine in more detail the
integrity of the Wnt/-catenin pathway in NPC tumours focussing on the expression of key positive
(Wnt/-catenin) and negative (APC, Axin,-TRCP, sFRP1, DKK, GSK3) regulators of the Wnt/catenin signalling pathway. The impact of EBV infection on Wnt/-catenin signalling will also be examined
in nasopharyngeal epithelial cell lines stably infected with EBV. Similarly, the impact of EBV infection on
the induction of an epithelial ‘stem cell phenotype” will also be explored, focusing on the contributions of
dysregulated Wnt/-catenin signalling to these effects.
Techniques to be used in the project:
Cell culture (Cell proliferation/apoptosis/sphere formation assays), immunohistochemical staining, indirect
immunofluorescence staining and confocal imaging, western blotting, RT-PCR, quantitative real-time RTPCR, Luciferase reporter assays, Fluorescence activated cell sorting (FACS).
Are these techniques already established in the laboratory where the project would take place?
Yes
Names of other researchers who may also be involved in project (whether supervisory or technical):
Miss Rebecca J.Port (PhD student)
Miss Sonia P.Maia (Research technician)
Dr John R. Arrand (Senior Research Fellow)
Key References:
(1). Young, L.S. and Rickinson, A.B. (2004). Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4:757768.
(2). Wang, J., Guo, L.P., Chen, L.Z., Zeng, Y.X. and Lu, S.H. (2007). Identification of cancer stem cell-like
side population cells in human nasopharyngeal carcinoma cell line.
Cancer Res. 67:3716-24.
(3) Katoh, M. (2007). Networking of WNT, FGF, Notch, BMP, and Hedgehog signaling pathways during
carcinogenesis. Stem Cell Reviews and Reports 3:30-38.
Does this project require the student to hold a personal home office licence?
No
Do you have all the relevant ethical approval for this study?
Yes
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Professor John Heath / Dr Josh Rappoport
Contact details: j.k.heath@bham.ac.uk
Office Location: Biosciences Tower 5th & Centre for System Biology
School: Biosciences
Investigating the interaction of key components of the in Fibroblast Growth Factor Signalling
pathway
Outline of Project:
The Fibroblast Growth Factor (FGF) family of ligands mediate a wide variety of cellular processes in
development, tissue repair and physiological homeostasis. Dys-regulation of the FGF pathway is a frequent
feature of tumour cells and is also central to a number of congenital diseases. Pathological manifestations of
FGF signalling involve changes in signalling dynamics consequent upon the actions of intracellular
pathways that regulate the trafficking of activated FGF receptors. A key component of the early phases of
FGF receptor trafficking is the requirement for the multifunctional signalling adaptor EPS8 to recruit
activated receptors from the cell surface and direct them to specific intercellular compartments. In studying
the protein partners of EPS8 we identified NBR1 – a multifunctional scaffold protein that we have
previously shown to be involved in receptor degradation and autophagy. We now wish to establish where in
the cell the EPS8/NBR1 interaction occurs and what the consequences of this interaction are for receptor
degradation and signalling. The former question will be addressed by live cell imaging using fluorescently
tagged versions of EPS8 and NBR1 and the latter questions addressed by RNAi mediated silencing of EPS8
and NBR1 and by studying the biological actions of mutant forms of EPS8 and NBR1 in mediating the FGF
response.
Techniques to be used in the project: Cell culture, live cell imaging, western blotting.
Are these techniques already established in the laboratory where the project would take place?
Yes
Names of other researchers who may also be involved in project (whether supervisory or technical):
Dr Debbie Cunningham
Key References:
Mardakheh FK, Yekezare M. Machesky LM, and Heath JK (2009) Spred regulates activated receptor trafficking to suppress
ERK1/2 via the novel late endosomal protein NBR1. J Cell Biol. 2009 Oct 19;187(2):265-77.
Mardakheh FK, Auciello G, Dafforn TR, Rappoport JZ, Heath JK. (2010) Nbr1 is a novel inhibitor of ligand-mediated RTK
degradation. Mol Cell Biol. 30, 5672-5685 PubMed PMID: 20937771
Sandilands E, Akbazardeh S, McEwan D, Frame M and Heath JK (2007) Src kinase dictates the activation, trafficking and
signalling dynamics of Fibroblast Growth Factor Receptors. EMBO Reports 12):1162-9.
Does this project require the student to hold a personal home office licence?
No
Do you have all the relevant ethical approval for this study?
Yes
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Prof Ann Logan & Dr Graham Wallace
Contact details: a.logan@bham.ac.uk
Office Location: IBR (west) 2nd floor
School: CEM/I&I
Glaucoma and Inflammation
Outline of Project:
Glaucoma is an inflammatory disease. The major risk factor for glaucoma and the focus of treatment is
increased intraocular pressure. Intraocular pressure is a function of production of liquid aqueous humor by
the ciliary processes of the eye and its drainage through the trabecular meshwork. Aqueous humor flows
from the ciliary processes into the posterior chamber, bounded posteriorly by the lens and anteriorly by the
iris. It then flows through the pupil of the iris into the anterior chamber, bounded posteriorly by the iris and
anteriorly by the cornea. From here the trabecular meshwork drains aqueous humor via Schlemm's canal
into scleral plexuses and the general blood circulation.
In open angle glaucoma there is reduced flow through the trabecular meshwork; in angle closure glaucoma,
the iris is pushed forward against the trabecular meshwork, blocking fluid from escaping. In both cases,
inflammatory cytokines, produced by in myeloid cells, that induce fibrotic scarring has been causatively
implicated, so that the drainage portals are blocked, resulting in raised intraocular pressure.
This project will use molecular, cellular and imaging methods in a new rat model of glaucoma and also in
samples from glaucoma patients to investigate the cells ie macrophages, and fibrogenic cytokines associated
with uveitis, trabecular meshwork scarring and raised intraocular pressure. It will also evaluate the potential
use of novel anti-inflammatory and neuroprotective agents in reducing inflammatory cascades, thereby
suppressing disease-related increases in intraocular pressure and retinal degeneration.
Techniques to be used in the project:
Ocular surgery; immunohistochemistry; genomics/proteomics/metabolomics; cell culture; flow cytometry
Names of other researchers who may also be involved in project (whether supervisory or technical):
Si Rauz (Supervisory)
Shabbir Mohamed (Supervisory)
Key References:
1. Danesh-Meyer HV. Neuroprotection in glaucoma: recent and future directions. Curr Opin Ophthalmol.
2011 Mar;22(2):78-86. Review.
2. Varma R, Peeples P, Walt JG, Bramley TJ. Disease progression and the need for neuroprotection in
glaucoma management. Am J Manag Care. 2008 Feb;14(1 Suppl):S15-9.
3. Rhee DJ, Haddadin RI, Kang MH, Oh DJ. Matricellular proteins in the trabecular meshwork. Exp Eye
Res. 2009 Apr;88(4):694-703
Does this project require the student to hold a personal home office licence?
Yes
Do you have all the relevant ethical approval for this study?
Yes
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Dr Heather Long and Prof Alan Rickinson
Contact details: h.m.long@bham.ac.uk 0121 4142808
a.b.rickinson@bham.ac.uk 0121 4144490
Office Location: Institute for Cancer Sciences room 419
School: School of Cancer Sciences
Therapeutic exploitation of CD4+ T cells - characterising the CD4+ T cells used therapeutically to
treat post-transplant lymphoproliferativedisease (PTLD).
Outline of Project:
It is now clear that in many settings CD4+ T cells can directly recognise and kill MHC class II-positive
target cells expressing their cognate antigen. For malignancies such as lymphoma occurring within
constitutively MHC II-expressing cells, this opens up exciting possibilities for therapeutic exploitation of
CD4+ T cells. In this context, Epstein-Barr virus (EBV)-associated post-transplant lymphoproliferative
disease (PTLD) can be cured by adoptive transfer of T cells generated by in vitro stimulation with EBVtransformed lymphoblastoid cell lines (virus-infected cells that resemble PTLD). In a recent study, PTLD
patients who receive infusions of 3rd party LCL-stimulated T cell preparations containing higher
percentages of CD4+ T cells achieved better long-term clinical responses. However, the CD4+ component
of the blood is phenotypically and functionally heterogeneous, and the identity and functional characteristics
of the CD4+ T cells present remains to be determined.
This project will use multi-colour flow cytometric analysis and our novel MHC class II tetramers to assess
the specificity and characteristics of the CD4+ T cells present in the LCL-stimulated preparations used
therapeutically. A better understanding of these crucial immune effectors and their relevant immune targets
may eventually lead to swifter preparation of therapeutic T cell lines against more clinically relevant
antigenic targets, and hence improve the future long-term survival of patients with EBV-associated B cell
malignancies.
Techniques to be used in the project:
Flow cytometry
Culture of T cell lines
Immunological assays of cytokine release
Are these techniques already established in the laboratory where the project would take place?
Yes/No
Names of other researchers who may also be involved in project (whether supervisory or technical):
Miss Alison Leese, Senior Research Technician
Key References:
Long HM, Leese AM, Chagoury OL, Connerty SR, Quarcoopome J, Quinn LL, Shannon-Lowe C, Rickinson AB. (2011)
Cytotoxic CD4+ T cell responses to Epstein-Barr virus contrast with CD8 responses in breadth of lytic cycle antigen choice and
in lytic cycle recognition. J Immunol. 187:92-101.
Haque, T., G. M. Wilkie, M. M. Jones, C. D. Higgins, G. Urquhart, P. Wingate, D. Burns, K. McAulay, M. Turner, C. Bellamy,
P. L. Amlot, D. Kelly, A. MacGilchrist, M. K. Gandhi, A. J. Swerdlow, and D. H. Crawford. 2007. Allogeneic cytotoxic T-cell
therapy for EBV-positive posttransplantation lymphoproliferative disease: results of a phase 2 multicenter clinical trial. Blood
110:1123-1131.
Does this project require the student to hold a personal home office licence?
Yes/No
Do you have all the relevant ethical approval for this study?
Yes/No
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Professor Chris McCabe and Professor Jayne Franklyn
Contact details: 58713; mccabcjz@bham.ac.uk
Office Location: IBR Floor 2, Office 223
School: CEM
PBF and genomic instability in the thyroid.
Outline of Project:
Pituitary tumor transforming gene (PTTG)-binding factor (PBF or PTTG1IP) is a little characterized protooncogene which we have shown to be implicated in breast, colon and thyroid tumourigenesis.1,2 We recently
determined that PBF overexpression results in thyroid hyperplasia and genetic instability in vivo, with a
parallel increase in expression of the DNA repair gene Rad6. In vitro, PBF repressed both p53 function and
stability. As PBF induces Rad6, and Rad6 is capable of altering the stability of p53, we now wish to unite
these novel findings, and determine whether PBF directly impairs DNA repair by promoting Rad6
dysregulation of p53.
Figure 1. Mice over-expressing PBF have enlarged thyroid glands
(left) with increased Rad6 expression (centre left), genetic
instability and hyperplasia (centre right). In K1 thyroid cells, PBF
potently reduces p53 stability (right).
The student would join the leading thyroid cancer research group in the UK, a successful and vibrant mix of
clinical and non-clinical scientists. Given the central importance of p53 to human cancer, this project has the
potential to yield genuinely novel and important data.
Techniques to be used in the project:
Transfection, siRNA knock-down, ubiquitination assays, cell culture, Western blotting, data analysis, critical appraisal
of literature, scientific writing.
Are these techniques already established in the laboratory where the project would take place?
Yes
Names of other researchers who may also be involved in project (whether supervisory or technical):
Dr Martin Read, Dr Vicki Smith, Mr Neil Sharma, Robert Seed, Gavin Ryan.
Key References:
1. Read ML, Lewy GD, Fong JC, Sharma N, Seed RI, Smith VE, Gentilin E, Warfield A, Eggo MC, Knauf JA,
Leadbeater WE, Watkinson JC, Franklyn JA, Boelaert K, McCabe CJ. (2011). Proto-oncogene PBF/PTTG1IP
regulates thyroid cell growth and represses radioiodide treatment. Cancer Research. 71(19):6153-64.
2. Watkins RJ, Read ML, Smith VE, Sharma N, Reynolds GM, Buckley L, Doig C, Campbell MJ, Lewy G, Eggo
MC, Loubiere LS, Franklyn JA, Boelaert K, McCabe CJ. (2010) Pituitary tumor transforming gene binding
factor: a new gene in breast cancer. Cancer Research. 70(9):3739-49.
Does this project require the student to hold a personal home office licence?
No
Do you have all the relevant ethical approval for this study?
Yes
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Professor Jane McKeating and Professor David Adams
Contact details: j.a.mckeating@bham.ac.uk
Office Location: Institute for Biomedical Research office 534
School: Immunity and Infection
In vitro modelling the cytomegalovirus and hepatitis C virus co-infected liver.
Outline of Project:
Chronic hepatitis C virus (HCV) related cirrhosis is the most common indication for liver transplantation in Europe
and the US. HCV infects the newly transplanted liver in all cases, however, disease recurrence varies with some
individuals showing rapid progression and others minimal disease1. Our current understanding of the role host or viral
parameters play in disease progression is limited and requires further investigation. Human cytomegalovirus (CMV)
is the most common cause of infection following liver transplantation and is a common cause of morbidity post liver
transplant. CMV infection not only causes direct effects in target organs (e.g., hepatitis), but has a number of indirect
effects, including a general immunosuppressive syndrome that has the potential to influence the recurrence and/or
severity of HCV replication2. Seckert and colleagues recently reported that liver sinusoidal endothelial cells are the
major site of murine CMV latency and reactivation in the liver 3. Observations in our laboratory demonstrating that
liver sinusoidal cells regulate HCV replication in adjacent hepatocytes by modulating bone morphogenic protein 4
(BMP4) expression4, provide an interface for these pathogens to interact and deregulate host immune responses.
Hypothesis: CMV infection of liver sinusoidal endothelial cells perturbs local inflammatory immune responses that
contribute to HCV associated liver disease.
Objectives: To study the effects of CMV infection on liver sinusoidal endothelial expression of inflammatory
mediators by PCR based microarray.
Techniques to be used in the project: The student will be trained in the preparation of cellular RNA and
quantitative real time PCR technologies to measure the abundance of mRNA species encoding a defined number of
inflammatory mediators. Initial studies will address the effect(s) of CMV on the primary endothelial cell
inflammasome and results will be validated with RNA preparations form infected liver tissue. These techniques will
provide a generic skill base that is widely applicable to a range of biologically diverse projects.
Are these techniques already established in the laboratory where the project would take place? Yes
Names of other researchers who may also be involved in project (whether supervisory or technical):
Dr Tony Burns a CMV expert and Drs Luke Meredith and Ian Rowe, hepatitis C virologists.
Key References:
1. Berenguer M. What determines the natural history of recurrent hepatitis C after liver transplantation? 2005. J Hepatol 42: 448456.
2. Mocarski ES. Immune escape and exploitation strategies of cytomegaloviruses: impact on and imitation of the major
histocompatibility system. 2004. Cell Microbiol 6: 707-717.
3. Seckert et al. Liver sinusoidal endothelial cells are a site of murine cytomegalovirus latency and reactivation. 2009. J Virol 83:
8869-84.
4. Rowe et al. Liver sinusoidal endothelial cells regulate hepatitis C virus replication through vascular endothelial growth factor
mediated suppression of bone morphogenetic protein 4. Submitted to J Clin Invest.
Does this project require the student to hold a personal home office licence? No
Do you have all the relevant ethical approval for this study? Yes
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Professor Jane McKeating and Dr Peter Balfe
Contact details: j.a.mckeating@bham.ac.uk
Office Location: Institute for Biomedical Research office 534
School: Immunity and Infection
A novel approach to the design of anti-viral peptides to inhibit hepatitis C virus infection
Outline of Project:
Hepatitis C virus (HCV) is a globally important pathogen with over 170 million infected individuals worldwide at
risk of developing progressive liver disease. Current treatments for chronic hepatitis C are limited and new antiviral agents are urgently needed. Pathogen entry into a host cell is defined by high affinity interaction(s) between
virus encoded proteins and cellular molecules or receptors that initiate one of the earliest steps in the viral
lifecycle1.
Recent advances have identified the essential role of tetraspanin CD81 and tight
junction protein claudin-1 receptor complexes in HCV entry2. Our laboratory has
modelled the molecular interface between CD81 and claudin-1 large extracellular
loops, allowing the identification of contact residues in both CD81 and claudin-1
that are essential for HCV entry3. We have modelled a series of peptides that will
disrupt the receptor complex and this project will evaluate the effect(s) of these
peptides on HCV infection and hepatoma cell biology. This data will increase
our understanding of the molecular mechanism of HCV entry and will help
elucidate a biological role for this receptor complex in hepatocyte biology.
Hypothesis: Peptide disruption of CD81 and claudin-1 receptor complexes will
inhibit HCV entry.
Objectives: To study the effects of recombinant peptides on HCV pseudoparticle
infection and hepatoma biology.
Techniques to be used in the project: The student will be trained in cell culture, virus propagation and high
resolution imaging of CD81 and claudin-1 trafficking, including Fluorescent Recovery After Photobleaching
(FRAP) and Fluoresecent Resonance Energy Transfer (FRET). These techniques provide a generic skill base that is
widely applicable to a range of biologically diverse projects. The student will join a vibrant laboratory of basic and
clinical scientists working on liver disease and will join weekly lab meeting and journal clubs.
Are these techniques already established in the laboratory where the project would take place? Yes
Names of other researchers who may also be involved in project (whether supervisory or technical):
Drs Nicola Fletcher and Zania Stamataki - hepatitis C virologists.
Key References:
1.
2.
3.
Thorley, J.A., McKeating, J.A. and Rappoport, J. (2010). Mechanisms of virus entry: sneaking in the front door.
Protoplasma, 244, 15-24.
Harris, H.J., Davis, C., Mullins J.G.L., Hu, K., Goodall, M., Balfe, P., and McKeating, J.A. (2010). Claudin association
with CD81 defines hepatitis C virus entry. J.Biol.Chem, 285, 21092-102.
Davis, C., Harris, H.J., Balfe, P., Mullins, J.G., and McKeating, J.A. Molecular modeling the claudin-1 and CD81 hepatitis
C virus receptor complex. Submitted to Nature Structural & Molecular Biology.
Does this project require the student to hold a personal home office licence? No
Do you have all the relevant ethical approval for this study? Yes
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Professor Paul Murray, Dr. Wenbin Wei
Contact details: murraypg@adf.bham.ac.uk, W.Wei@bham.ac.uk
Office Location:
School: Cancer Sciences
Title of project: Investigation of genetic changes associated with poor outcome paediatric Hodgkin’s
lymphoma
Outline of Project:
Hypothesis and aims: We hypothesize that genetic changes not only drive Hodgkin’s lymphoma pathogenesis but
will also define clinically relevant subgroups of patients for stratified medicine [1, 2]. The aim of this project is to
identify and characterize the genetic changes associated with poor outcome Hodgkin’s lymphoma patients and to
investigate if existing drugs not previously used in the treatment of Hodgkin’s lymphoma could be used for patients
carrying particular mutations.
Background: A subset of children with Hodgkin’s lymphoma has disease that is either refractory to treatment or
which relapses early; outcome for these groups is particularly poor. Moreover, the level of chemosensitivity as
measured by early response to treatment is a prognostic feature without a known biological basis. There are no
studies to date which have addressed the underlying biology of the malignant cell population in these patients. High
throughput genome-wide sequencing is now used widely to detect mutations that drive oncogenesis [3, 4]. For
example, the International Cancer Genome Consortium (ICGC) is sequencing 50 different tumour types and/or
subtypes (http://www.icgc.org/). Because the malignant Hodgkin/Reed-Sternberg (HRS) cells make up less than 1%
of the total tumour mass, it has not been previously possible to sequence DNA from these cells using standard
sequencing protocols. However, it has recently been shown that whole genome sequencing can be performed on
single human cells, providing for the first time the opportunity to identify the global genetic changes associated with
Hodgkin’s lymphoma [5]. As part of our on-going omics-driven research programme in Hodgkin’s lymphoma [6, 7],
this project will apply single cell whole genome amplification technology to the investigation of genetic changes
associated with poor outcome paediatric Hodgkin’s lymphoma.
Plan of investigation:
1) Search for known mutations in Hodgkin’s lymphoma using “the Catalogue of Somatic Mutations in Cancer”
[4].
2) Search drugbank and the NCI drug development databases [8, 9] to determine if these mutated genes are known
drug targets.
3) Sequence the Hodgkin’s lymphoma cell line, L1236, using both unamplified DNA and amplified DNA from
laser-micro-dissected cells to assess the error rate introduced by laser-micro-dissection and amplification. DNA
will be amplified first using GenomePlex® Single Cell Whole Genome Amplification Kit and further with PCR.
4) Sequence three more Hodgkin’s lymphoma cell lines (L428, L591 and KMH2) to identify recurrent mutations
that can be used to test gene specific drug effects on growth inhibition and cytotoxicity.
5) Perform laser micro-dissection of HRS cells from patients. Amplify the DNA for Sanger sequencing to establish
the frequencies of known mutations in both good and poor outcome patients.
Techniques to be used in the project:
Laser micro-dissection, Amplification of DNA using GenomePlex® Single Cell Whole Genome
Amplification Kit, PCR, Sanger sequencing and sequence analysis, Use of public bioinformatics databases
Are these techniques already established in the laboratory where the project would take place?
Yes
Names of other researchers who may also be involved in project (whether supervisory or technical):
Dr. Pamela Kearns and Dr. John Arrand
Key References:
1. Stratified medicine http://en.wikipedia.org/wiki/Stratified_medicine
2. Targeted therapy http://en.wikipedia.org/wiki/Targeted_therapy
3. The Cancer Genome Atlas Research Network (2011). Integrated genomic analyses of ovarian carcinoma.
Nature 474: 609–615.
4. Forbes et al. (2011) COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations
in Cancer. Nucl. Acids Res. 39 (suppl 1): D945-D950.
5. Navin et al. (2011) Tumour evolution inferred by single-cell sequencing. Nature 472: 90–94
6. Vrzalikova, K., M. Vockerodt, S. Leonard, A. Bell, W. Wei, A. Schrader, K. L. Wright, D. Kube, M. Rowe,
C. B. Woodman and P. G. Murray (2011). "Down-regulation of BLIMP1{alpha} by the EBV oncogene,
LMP-1, disrupts the plasma cell differentiation program and prevents viral replication in B cells: implications
for the pathogenesis of EBV-associated B-cell lymphomas." Blood 117(22): 5907-5917.
7. Murray, P. G., Y. Fan, G. Davies, J. Ying, H. Geng, K. M. Ng, H. Li, Z. Gao, W. Wei, S. Bose, J. Anderton,
G. Kapatai, G. Reynolds, A. Ito, T. Marafioti, C. B. Woodman, R. Ambinder and Q. Tao (2010). "Epigenetic
Silencing of a Proapoptotic Cell Adhesion Molecule, the Immunoglobulin Superfamily Member IGSF4, by
Promoter CpG Methylation Protects Hodgkin Lymphoma Cells from Apoptosis." Am J Pathol 177: 14801490.
8. Knox et al. DrugBank 3.0: a comprehensive resource for 'omics' research on drugs. Nucleic Acids Res.
2011 Jan;39(Database issue):D1035-41.
9. Lu, X., W. Wei, J. Fenton, M. S. Nahorski, E. Rabai, A. Reiman, L. Seabra, Z. Nagy, F. Latif and E. R.
Maher (2011). Therapeutic targeting the loss of the birt-hogg-dube suppressor gene. Mol Cancer Ther
10(1): 80-9.
Does this project require the student to hold a personal home office licence?
Do you have all the relevant ethical approval for this study?
We expect to have all the relevant ethical approval before this project starts.
No
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Professor Gerard Nash & Dr. Helen McGettrick
Contact details: g.nash@bham.ac.uk (x43670); h.m.mcgettrick@bham.ac.uk
(x44043)
Office Location: N119, N118; 1st Floor IBR West, The Medical School
School: Clinical and Experimental Medicine; Immunity and Infection
Novel regulatory steps in migration of T-cells and neutrophils into and out of tissue during
inflammation
Outline of Project:
In inflammation, circulating leukocytes are recruited by blood vascular endothelium (VEC), and
migrate into the tissue where they fulfil their functions in the destruction of invading pathogens and
remodelling of damaged tissue. Once the trigger has been eliminated, recruited cells must be cleared to
allow resolution. Neutrophils undergo apoptosis and are phagocytosed locally by macrophages, while Tcells migrate across the lymphatic endothelium (LEC) to exit the tissue and return to the circulation, via
lymph nodes. How leukocytes enter tissue is well documented (e.g.1-3), but little is known about the
regulation of migration through LEC and the basis of selectivity between types of leukocytes. Using multicellular in vitro models, we have shown that tissue cells (fibroblasts) can regulate the recruitment of
leukocytes to vascular endothelium and their subsequent migration (4,5). We hypothesise that LEC can also
provide signals that modify leukocyte migration through blood vascular endothelium, and importantly,
allow T-cell migration out of the tissue, whilst stopping neutrophils.
This project will use a novel multi-cellular migration assay, which will enable us to compare the
ability of neutrophils and lymphocytes to migrate through activated VEC and subsequently LEC. The main
readouts will be the levels of neutrophil and lymphocyte migration through the different compartments. We
will also determine the phenotype of lymphocytes migrating across the different layers (e.g. memory vs.
naive, T-cell subsets, B-cells, NK-cells), to test for selectivity between these populations. We aim to gain
insight into the processes regulating leukocyte clearance from tissue, and to examine the concept that
encouraging exit could be a strategy to resolve chronic inflammatory conditions.
Techniques to be used in the project: Cell culture, isolation of T-cells, adhesion and migration assays,
image analysis, immunofluorescence and flow cytometry
Are these techniques already established in the laboratory where the project would take place?
Yes/No
Names of other researchers who may also be involved in project (whether supervisory or technical):
This project is part of a collaboration between the supervisors and Dr. Ed Rainger and Prof. Chris Buckley
Key References:
1. Springer, 1995. Annual Reviews in Physiology. 57: 827-872
2.Marelli-Berg et al., 2008, Journal of Pathology. 214: 179–189
3. Nourshargh et al., 2010. Nature Reviews (molec cell biol) 11: 366-378
5. McGettrick et al., 2010 European Journal of Immunology. 39: 98-107
6. McGettrick et al., 2011 Immunology. 131: 357-370
Does this project require the student to hold a personal home office licence?
Yes/No
Do you have all the relevant ethical approval for this study?
Yes/No
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Professor Michael Overduin; Dr Martyn Chidgey
Contact details: M.Overduin@bham.ac.uk; M.A.Chidgey@bham.ac.uk
Office Location: Cancer Studies Bldg; Clinical Research Block
School: Cancer Sciences
Determination of the effect of inherited mutations on the structure and function of the desmosomal
protein plakoglobin (γ catenin).
Outline of Project:
Desmosomes are complex multi-protein intercellular junctions. They are located at the cell membrane
where they rivet cells together. They are essential for the maintenance of tissue integrity. Loss of
desmosomal function in cardiomyocytes results in arrhythmogenic right ventricular cardiomyopathy
(ARVC), one of the most common inherited cardiomyopathies, and a cause of heart failure and sudden
death in young adults, particularly competitive athletes. Loss of desmosomal adhesion in the epidermis
causes lethal acantholytic epidermolysis bullosa, a distressing skin blistering condition that results in
catastrophic fluid loss and infant death. Finally, loss of desmosomal adhesion has been implicated in
cancer. The aim of this project will be to determine the effect of inherited mutations on the structure and
function of the desmosomal protein plakoglobin (γ catenin). Wild-type and ARVC mutant plakoglobin
proteins will be expressed in bacteria and purified using established techniques. A variety of biophysical
techniques will then be used to investigate the effects of the mutations on plakoglobin structure. The ability
of the wild-type and mutant plakoglobin proteins to interact with the plakoglobin ligands desmocollin and
desmoglein will be compared.
Techniques to be used in the project:
Bacterial expression, protein purification, size exclusion chromatography, circular dichroism, pull-down
assays, SDS-PAGE, Western blotting.
Are these techniques already established in the laboratory where the project would take place?
Yes
Names of other researchers who may also be involved in project (whether supervisory or technical):
Caezar Al-Jassar
Key References:
Al-Jassar, C., Knowles, T., Jeeves, M., Kami, K., Behr, E., Bikker, H., Overduin, M. and Chidgey, M. (2011).
The non-linear structure of the desmoplakin plakin domain and the effects of cardiomyopathy-linked mutations.
J. Mol. Biol. 411, 1049-1061.
Kami, K., Chidgey, M., Dafforn, T. and Overduin, M. (2009). The desmoglein-specific cytoplasmic region is
intrinsically disordered in solution and interacts with multiple desmosomal protein partners. J. Mol. Biol. 386,
531-543.
Garrod, D. and Chidgey, M. (2008). Desmosome structure, composition and function. Biochim. Biophys. Acta.
1778, 572-587.
Does this project require the student to hold a personal home office licence?
No
Do you have all the relevant ethical approval for this study?
Yes
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Dr. S Naeem Shafqat and Professor Wiebke Arlt
Contact details: Center for Endocrinology, Diabetes and Metabolism (CEDAM)
Email: S.N.Shafqat@bham.ac.uk tel office: 0121 4142764
Office Location: CEDAM (2nd floor IBR)
School: School of Clinical and Experimental Medicine
“Protein expression-purification profiling of P450 oxidoreductase disease mutants”
Back ground: Cytochrome P450 oxidoreductase (POR) is a flavoprotein that transfer electrons from
NADPH to all microsomal (type 11) Cytochrome P450 enzymes, which are responsible for metabolizing
majority of clinically used drugs and play crucial part in steroid biosynthesis.
P450 oxidoreductase (POR) deficiency: A rare autosomal recessive disorder, caused by mutations in POR
gene, exhibiting severe deleterious effects on steroid biosynthesis, and on the ability of Cytochrome P450
enzymes to metabolize drugs.
Clinical Implications of POR mutations: The reported clinical symptoms in patients having POR
mutations includes skeletal malformations commonly referred as Antley-Bixler syndrome (ABS), and
disorders of sex developments (ambiguous genitalia, virilisation, premature puberty, girls born with testis or
boys with ovaries and uterus).
Biochemical investigation: To understand the deleterious effects of POR mutants, one has to study their
abnormal expression and functional profiles responsible for causing different clinical complications in
patients. These POR mutants will be subjected to vigorous protein expression-purification trials in order to
identify the best possible condition to express them in soluble form. The produced protein will later be
available to be used in functional assays, protein crystallization and structural studies.
Techniques to be used in the project:
-Gene primer design, Gene amplification using PCR, Advance cloning technique (Ligation independent cloning),
E.coli transformation, Protein expression optimization in E.coli, Protein purification, Protein production for the
purpose of crystallization, functional assays and 3D-structure determination.
Are these techniques already established in the laboratory where the project would take place?
Yes
Names of other researchers who may also be involved in project (whether supervisory or technical):
None
Key References:
1) Arlt W, et al. (2004). “Congenital adrenal hyperplasia caused by mutant P450 oxidoreductase and
human androgen synthesis: analytical study” Lancet 2004; 363: 2128–35.
2) Miller WL, Agarwal V, Sandee D, et al. (2011). "Consequences of POR mutations and
polymorphisms." Molecular and Cellular Endocrinology 336 (2011) 174–179.
3) Huang N, et al (2005). “Diversity and Function of Mutations in P450 Oxidoreductase in Patients
with Antley-Bixler Syndrome and Disordered Steroidogenesis.” Am. J. Hum. Genet. 76:729–749.
Does this project require the student to hold a personal home office licence?
No
Do you have all the relevant ethical approval for this study?
Yes
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors:
1) Alex Sinclair
2) Jeremy Tomlinson
Contact details:
1) a.b.sinclair@bham.ac.uk
2) j.w.tomlinson@bham.ac.uk
Office Location:
1) IBR West Extension
2) IBR floor 2
1
2
School: Neurobiology and Neuropharmacology and Centre for Endocrinology, Diabetes &
Metabolism, School of Clinical and Experimental Medicine, College of Medical and Dental Sciences
Investigating the potential of 5α-reductase inhibition in modifying CSF secretion- possibilities for
therapeutic use in idiopathic intracranial hypertension.
Outline of Project:
Idiopathic intracranial hypertension (IIH) is a condition of unknown aetiology, characterised by elevated
intracranial pressure in young obese women which leads to blindness. Our previous work has highlighted a
potential role of 5α-reductase (a cortisol inactivating enzyme with an additional key role in activating
testosterone to 5α dihydrotestosterone). We demonstrated that amongst patients with IIH, 5α-reductase
activity declined following therapeutic weight loss. Changes in androgen metabolism may have important
implications in the female biased condition of IIH. Additionally, 5α-reductase is know to be regulated by
oestrogen. Our initial findings suggest that 5α-reductase may be important in IIH. The mechanism
underlying the role of 5α-reductase in IIH and intracranial pressure regulation now needs to be explored.
Intracranial pressure regulation depends on the balance between production of cerebrospinal fluid (CSF) by
the choroid plexus (CP) and drainage at the arachnoid granulation tissue (AGT). We have localized 5αreductase type 1 to our CP and AG primary epithelial cell line.
We propose quantifying levels of 5α-reductase expression (as well a key glucocorticoid target genes and ion
channels central to fluid secretion) in CP and AG cell cultures in the presence and absence of a specific 5αreductase inhibitor using real time PCR and ELISA’s. A model of CSF secretion will be established using a
monolayer of CP cells grown on a Transwell filter. Changes in fluid distribution above and below the filter
will be monitored following treatment with 5α-reductase inhibitors. This cutting edge work is likely to
contribute to a peer-reviewed scientific publication.
Techniques to be used in the project: Cell culture with transfection and gene over expression and gene
silencing (SiRNA), enzyme assays (with chemical inhibitor studies), RNA extraction, PCR, real time PCR,
low density PCR arrays (potentially), ELISA and Western Blot analyais, transepithelial electrical resistance
monitoring and Transwell culture systems.
Are these techniques already established in the laboratory where the project would take place?
Yes
Names of other researchers who may also be involved in project (whether supervisory or technical):
Technical support from Laura Gathercole (Postdoctoral Research Fellow)
Key References:
Sinclair AJ et al. Low calorie diet and intracranial pressure in idiopathic intracranial hypertension: a prospective cohort
study . BMJ. 2010 Jul 7;341:c2701.
Sinclair AJ et al. Cerebrospinal fluid corticosteroid levels and cortisol metabolism in patients with idiopathic intracranial
hypertension: a link between 11beta-HSD1 and intracranial pressure regulation? J Clin Endocrinol Metab. 2010
Dec;95(12):5348-56. Epub 2010 Sep 8.
Haselbach M et al. Porcine choroid plexus epithelial cells in culture: Regulation of barrier properties and transport
processes. Microscopy Research and Technique 2001 52 (1): 137-152.
Vassiliadi DA et al. Increased 5 alpha-reductase activity and adrenocortical drive in women with polycystic ovary
syndrome. J Clin Endocrinol Metab. 2009 Sep;94(9):3558-66. Epub 2009 Jun 30.
Does this project require the student to hold a personal home office licence?
Yes/No
Do you have all the relevant ethical approval for this study?
Yes/No
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Dr Zania Stamataki, Professor Jane McKeating
Contact details: z.stamataki@bham.ac.uk 01214158673
Office Location: 5th Floor IBR Building, Medical School.
School: Immunity and Infection
Project Title: T cell entosis in the liver: It’s a cell-eat-cell world.
Outline of Project:
Entosis is the process whereby one cell invades another, and can remain internalised in its own vesicle in the
host cell’s cytoplasm for long periods of time. The internalised cell may die or it may be released unharmed,
and little is known regarding the molecules guiding these decisions. We recently discovered that T cells
internalise into hepatocytes in the liver via entosis. This may have implications for the role of T cells in the
regulation of liver immunity, and it is therefore a hot topic in immunology as well as cell biology. The
successful candidate will learn how to culture human hepatocyte and lymphocyte cell lines, label them with
fluorescent markers and acquire confocal microscopy images of internalised cells. Internalised and released
T cells will be quantified by flow cytometry. Lymphocyte entosis will be performed in the presence of a
series of chemical inhibitors for endocytic pathways, to help identify the molecules involved in
lymphocyte entosis into hepatocytes. Entosis will also be performed in the presence of pro-inflammatory
cytokines, to monitor T cell release from hepatocytes. The techniques used have already been established
in our lab and the results will form part of a scientific publication currently in preparation on the subject.
The successful candidate will benefit from the opportunity to work in a vibrant laboratory with clinical and
academic scientists, and learn advanced techniques in liver biology, immunology and cell biology, with a
special focus on chronic inflammation and viral infection.
Techniques to be used in the project:
Cell culture, labelling cells with fluorescent markers and antibodies, flow cytometry, confocal microscopy,
T cell and liver immunology, cell biology of cell-in-cell structures and intracellular vesicles.
Are these techniques already established in the laboratory where the project would take place?
Yes/No
Names of other researchers who may also be involved in project (whether supervisory or technical):
Professor David Adams, Dr Peter Balfe.
Key References:
1.
Overholtzer, M. and J.S. Brugge, The cell biology of cell-in-cell structures. Nat Rev Mol Cell Biol,
2008. 9(10): p. 796-809.
2.
Overholtzer, M., et al., A nonapoptotic cell death process, entosis, that occurs by cell-in-cell
invasion. Cell, 2007. 131(5): p. 966-79.
3.
Benseler, V., et al., Hepatocyte entry leads to degradation of autoreactive CD8 T cells. Proc Natl
Acad Sci U S A, 2011. 108(40): p. 16735-40.
Does this project require the student to hold a personal home office licence?
Yes/No
Do you have all the relevant ethical approval for this study?
Yes/No
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Dr Kai-Michael Toellner
Contact details: K.M.Toellner@bham.ac.uk
Office Location: IBR, room 432
School: Immunity and Infection
The role of IgG for germinal centre B cell selection?
Outline of Project:
Germinal centres (GCs) are the sites of B cell affinity maturation through Darwinian evolution, comprising
B cell proliferation, hypermutation and selection (MacLenann 1994). It is not clear how antigen-specific
selection of high affinity B cell variants is achieved. We recently showed that soluble antibodies have a
major role in regulating selection through antigen shielding, producing a feedback loop that increases the
threshold for B cell selection dependent on the affinity of GC derived plasma cells (manuscript submitted).
So far, we have shown that IgM given during a GC reaction leads to selection of higher affinity B cell
variants. IgG has different avidity and effector functions to IgM. The project will test, how class switched
antibodies influence GC B cell selection. We will test whether IgG influences affinity maturation by
detecting specific antibodies in serum samples mice from treated with specific IgG during the GC response.
Further, spleens will be studied for germinal centre B cell differentiation using immunohistology. Cytokine
and chemokine production will be tested using RT-PCR from the same tissues. Depending on progress of
the project GC B and T cells will be sorted to detect expression of cytokines and costimulatory molecules.
This should give a picture on how IgG regulates GC B cell selection, and whether IgG may be useful as a
tool to manipulate responses to pathogens or vaccines.
Techniques to be used in the project:
Techniques used will be immunoenzymatic histology, stereological analysis of tissue sections, ELISA, real
time RT-PCR. Depending on progress, more detailed analysis of expression of GC B cell associated
differentiation and signalling molecules using fluorescence microscopy.
Are these techniques already established in the laboratory where the project would take place?
Yes/No
Names of other researchers who may also be involved in project (whether supervisory or technical):
Dr. Yang Zhang, Dr. Jennifer Marshall
Key References:
MACLENNAN, I. C. M. 1994. Germinal centers. Annual Review of Immunology, 12, 117-139.
MEYER-HERMANN, M., FIGGE, M. T. & TOELLNER, K.-M. 2009. Germinal centres seen through the
mathematical eye: B cell models on the catwalk. Trends Immunol, 30, 157-164.
VICTORA, G. D., SCHWICKERT, T. A., FOOKSMAN, D. R., KAMPHORST, A. O., MEYER-HERMANN, M.,
DUSTIN, M. L. & NUSSENZWEIG, M. C. 2010. Germinal Center Dynamics Revealed by Multiphoton Microscopy
with a Photoactivatable Fluorescent Reporter. Cell, 143, 592-605.
Does this project require the student to hold a personal home office licence?
Yes/No
Do you have all the relevant ethical approval for this study?
Yes/No
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors:
Contact details:
Office Location:
School:
Professor Bryan Turner, Dr Ferenc Mueller and Professor
Laura O’Neill
b.m.turner@bham.ac.uk
IBR 331
Immunity and Infection, CEM
Investigating the effect of environmental toxins on histone modifications across developmentally
regulated genes.
Outline of Project:
Embryonic development is a dynamic process in which a terminally differentiated cell, the oocyte,
transforms into a pluripotent and essentially undifferentiated cell mass that then begins a programme of
differentiation. These events involve targeted gene activation and repression driven by epigenetic
mechanisms that switch cells between undifferentiated (possibly stem-cell like) and differentiated states.
Despite its fundamental importance, the interplay between epigenetic regulation and transcriptional control
remains mysterious. This project seeks to define epigenetic marks at the very earliest stages of embryo
development, how they are distributed across the genome and how they respond to environmental agents.
Such studies are technically challenging, largely because so few cells are available. We have developed a
novel chromatin immunoprecipitation (ChIP) protocol (Carrier ChIP, CChIP) that allows analysis of
chromatin from very small numbers of cells (1) and have used it to provide evidence for the heritability of
induced epigenetic change (2). CChIP can be applied to early zebrafish embryos (our unpublished results),
providing a powerful model system in which to study progressive epigenetic changes in a vertebrate embryo
(3). This project will map histone modifications across developmentally regulated genes, with and without
exposure to environmental toxins, and link these to phenotypic changes through development.
Techniques to be used in the project:
Culture of zebrafish embryos with and without environmental toxins, assessment of induced morphological
changes, harvesting of embryos and preparation of chromatin, Carrier Chromatin Immunoprecipitation
(CChIP) with antisera to modified histones, PCR.
Are these techniques already established in the laboratory where the project would take place?
Yes
Names of other researchers who may also be involved in project (whether supervisory or technical):
Dr Nan Li
Key References:
(1)
(2)
(3)
O’Neill LP et al (2006) Nature Genetics 38, 835-841
VerMilyea MD et al (2009) PLoS ONE 4 (6) e6086
Ferg et al., (2007) EMBO J. 26(17) 3945-56.
Does this project require the student to hold a personal home office licence?
No
Do you have all the relevant ethical approval for this study?
Yes
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Dr Graham Wallace; Dr Steve Young
Contact details: g.r.wallace@bham.ac.uk; s.p.young@bham.ac.uk
Office Location: (GW) Centre for Translational Inflammation Research. 1st floor
QE Hospital; (SY) 3rd floor IBR
School: Immunity and Infection
The effect of hormones on the Characteristics of macrophages
Outline of Project:
Monocytes are heterogeneous, but are defined in humans as CD11b+, CD11c+ and CD14+. In transfer
experiments CCR2+ monocytes are short-lived in naïve recipients, but migrated to inflamed sites where the
upregulate MHC class II. By contrast CX3CR1+ monocytes were found to persist after transfer and could be
recovered from the blood, spleen lungs, liver and brain. In vitro generated M1 macrophages, are easily
activated to produce NO when stimulated with LPS or IFN. LPS stimulation of M2 macrophages induces
arginase convertion of arginine to ornithine. Therefore these two populations of macrophages are very
different. Recent data has shown that epinephrine and 2 adrenoreceptor agonists potently inhibited LPSinduced TNF- and IL-12, but stimulated IL-10 production. The order of potency for hormones able to
inhibit IL-12 production ex vivo was: epinephrine > norepinephrine > 1,25-(OH)2 vitamin D3 >
hydrocortisone. This indicates that baseline epinephrine conditions cytokine responsiveness and through
this mechanism intrinsic hypo- or hyperactive adrenal medullas in some individuals may shape opposite
cytokine profiles. We have an interest in how these hormones control immune responses in our laboratory.
In this project M1 and M2 macrophages will be grown from human blood monocytes and the effects of the
above hormones will be tested to determine changes in profiles and response.
Techniques to be used in the project:
Cell culture, ELISA, PCR
Are these techniques already established in the laboratory where the project would take place?
Yes
Names of other researchers who may also be involved in project (whether supervisory or technical):
Key References:
Elenkov IJ. et al J Immunol 2008 181; 1737
Anane LH Brain Behav Immun. 2009 6:823-9.
Does this project require the student to hold a personal home office licence?
No
Do you have all the relevant ethical approval for this study?
Yes
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Dr Stephen Young
Contact details:s.p.young@bham.ac.uk. Tel: 0121 414 6480
Office Location: IBR 3rd floor room 332
School: Immunity & Infection
Immune cells as “farmers” of the stroma
Outline of Project:
Solid tumours are very low in oxygen and tumour cells may adapt to this environment by switching to
glycolytic energy production even when oxygen is available, the “Warburg effect” [1]. Tumour cells may
also secrete hydrogen peroxide which leads to the loss of mitochondria in the surrounding stromal
fibroblasts through autophagy [2]. With fewer mitochondria, these cells have to switch to glycolysis to make
ATP and, as a result, they secrete high energy metabolites such as lactate. The tumour cells may “harvest”
these metabolites to promote their growth, overcoming the hypoxic conditions in the tumour.
We hypothesize that, since most immune cells can produce hydrogen peroxide, this process may also occur
in “normal” inflammation, so that when immune cells migrate into hypoxic tissues to clear an infection,
they are able to generate metabolites from the surrounding stromal cells to allow their continued function. In
diseases such as rheumatoid arthritis this process may promote chronicity. We will test this hypothesis by
assessing the effects of oxygen manipulation and exposure to hydrogen peroxide and TNF on mitochondrial
numbers in cell lines (staining with mitotracker green). We will also assess markers of autophagy and relate
this to changes in metabolism, by assessing metabolic profiles using NMR spectroscopy. Cells will be
grown for various times in the inflammatory mediators (and strongly oxidising cigarette smoke extract –
since smoking is linked to rheumatoid arthritis [3]) to determine both acute and chronic effects on
mitochondria and metabolism. We will initially make use of cell lines to establish protocols and then use
primary blood cells to consolidate the results. If time allows we will also use primary stromal fibroblasts
from synovial tissue of rheumatoid arthritis patients.
Techniques to be used in the project: Tissue culture, flow cytometry, confocal microscopy, NMR
spectroscopy, biochemical analysis of cell metabolism
Are these techniques already established in the laboratory where the project would take place?
IBR labs and the Centre for Translation Research in Inflammation
Most are.
Names of other researchers who may also be involved in project (whether supervisory or technical):
Beth Clay, Martin Fitzpatrick, Rachel Bayley, Sabrina Kapoor, Karim Raza
Key References:
1.
Warburg O. On respiratory impairment in cancer cells. Science. 1956 Aug 10;124(3215):269-70.
2.
Martinez-Outschoorn UE, Lin Z, Trimmer C, Flomenberg N, Wang C, Pavlides S, et al. Cancer cells metabolically
"fertilize" the tumor microenvironment with hydrogen peroxide, driving the Warburg effect: Implications for PET imaging of
human tumors. Cell Cycle. 2011;10(15):2504-20.
3.
Kallberg H, Ding B, Padyukov L, Bengtsson C, Rannelid J, Klareskog L, et al. Smoking is a major preventable risk
factor for rheumatoid arthritis: estimations of risks after various exposures to cigarette smoke. Ann Rheum Dis. 2011;70(3):50811
Does this project require the student to hold a personal home office licence?
No
Do you have all the relevant ethical approval for this study?
Yes
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Dr Stephen Young
Contact details:s.p.young@bham.ac.uk. Tel: 0121 414 6480
Office Location: IBR 3rd floor room 332
School: Immunity & Infection
What is the role of the Lyp phosphatase (PTPN22) in phagocyte signalling?
Outline of Project:
A single nucleotide polymorphism in the PTPN22 gene is strongly associated with a number of autoimmune
diseases [1] and gives rise to a form of the Lyp phosphatase that fails to interact with the kinase csk [2].
This disrupts the regulation of the key TCR signalling kinase Lck and so may alter the process of negative
selection of autoreactive T cells in the thymus [1]. This variant also has effects in mature lymphocytes in the
form of poor or depressed signalling through the TCR and BCR [3].
However, the Lyp protein is expressed at the highest levels in phagocytes and at only modest levels in
lymphocytes [6]. The synovial membrane in rheumatoid arthritis, like most inflammatory sites, contains
large numbers of macrophages, which contribute to joint damage. It is possible then that the dysfunction of
the variant Lyp in phagocytes makes a substantial contribution to tissue damage in these diseases by
promoting dysfunction of the phagocytes. However, the function of Lyp in these cells has not been studied
and so it is not possible to suggest what the effects of the variant might be.
The student will test the hypothesis that PTPN22 regulates phagocyte activation and adhesion.
We will knock down Lyp expression in HL60, THP1 and U937 cell lines and study their responses to FcR
and TLR ligation.
We will transfect shRNA to knockdown Lyp expression. By using a viral plasmid construct (Qiagen
SABiosciences) permanent knockdown should be achievable. Lyp phosphatase activity will be assessed
using an assay we developed for CD45 [7]. Levels of PTPN22 protein will be monitored using western
blotting and mRNA using RT-PCR. Calcium signalling in response to TLR and FcR ligation will be
assessed along with production of superoxide (through cytochrome C reduction). Adhesion of the depleted
HL60 to substrates in a static adhesion assay will also be assessed. Whole cell phosphotyrosine
immunoblots, before and after activation, will reveal if substantial changes in phosphorylation are observed
and possibly identify specific hyperphosphorylated targets of Lyp in phagocytes.
Techniques to be used in the project: Tissue culture, flow cytometry, shRNA, biochemical analysis of cell
metabolism, adhesion assays
Are these techniques already established in the laboratory where the project would take place?
IBR labs and the Centre for Translation Research in Inflammation
Most are.
Names of other researchers who may also be involved in project (whether supervisory or technical):
Beth Clay, Martin Fitzpatrick, Rachel Bayley, Sabrina Kapoor, Karim Raza
Key References:
1. Vang T, Miletic AV, Bottini N, Mustelin T. Protein tyrosine phosphatase PTPN22 in human autoimmunity. Autoimmunity. 2007;40(6):45361.
2 .Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, et al. A functional variant of lymphoid tyrosine phosphatase is
associated with type I diabetes. Nat Genet. 2004;36(4):337-8.
3. Rieck M, Arechiga A, Onengut-Gumuscu S, Greenbaum C, Concannon P, Buckner JH. Genetic variation in PTPN22 corresponds to altered
function of T and B lymphocytes. J Immunol. 2007;179(7):4704-10.
4. Chien WW, Tidow N, Williamson EA, Shih LY, Krug U, Kettenbach A, et al. Characterization of a myeloid tyrosine phosphatase, Lyp, and
its role in the Bcr-Abl signal transduction pathway. J Biol Chem. 2003;278(30):27413-20.
5. Rider DA, Young SP. Measuring the specific activity of the CD45 protein tyrosine phosphatase. J Immunol Methods. 2003;277(1-2):125-32.
Does this project require the student to hold a personal home office licence?
No
Do you have all the relevant ethical approval for this study?
Yes
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Professor Steve P Watson
Contact details: s.p.watson@bham.ac.uk; tel: 4146514
Office Location: level 1: Institute of Biomedical Research
School: School: Clinical and Experimental Medicine
Regulation of shedding and internalisation of the FcRIIA receptor and the collagen receptor GPVI
Outline of Project:
The low affinity immune receptor, FcRIIA, and collagen receptor, GPVI-FcR -chain, signal through the
sequential activation of Src and Syk family kinases leading to activation of phospholipase C2 (PLC2). A key
feature of signalling by both receptors is a critical role for an immunoreceptor tyrosine-based activation motif
(ITAM), which is also used by other Fc receptors and by antigen receptors (e.g. T cell receptor). Signalling by
both receptors is regulated by receptor internalisation and by receptor shedding. This project will compare the
regulation of these two processes using a transfected cell line model in which the activation of PLC2 is
monitored using a reporter assay. The time course of activation of phospholipase C, receptor internalisation
(monitored by flow cytometry and by confocal microscopy using GFP tagged receptors) and receptor shedding
will be monitored. Experiments will be extended to cells deficient in various signalling proteins to further dissect
between these processes. All of the techniques are established in the group and training is available. The project
could form the base of a PhD programme.
Techniques to be used in the project: Protein biochemistry e.g. western blotting, immunprecipitation; cell
culture; reporter assays; flow cytometry; confocal and TIRF microscopy; molecular cloning
Are these techniques already established in the laboratory where the project would take place?
yes
Names of other researchers who may also be involved in project (whether supervisory or technical):
Craig Hughes and Alice Pollitt, Postdoctoral Fellows
Key References:
The reference shown below summarises work from the group on GPVI and a related receptor, CLEC-2, that could
also be investigated as a part of a PhD project
Watson, S.P., Herbert, J.M.J. and Pollitt, A.Y. (2010) Glycoprotein VI and CLEC-2 in hemostasis and vascular
integrity. J. Thromb. Haemost. 8, 1456-1467
Does this project require the student to hold a personal home office licence?
No
Do you have all the relevant ethical approval for this study?
Yes
Wellcome Trust Combined Training Programme
Short Laboratory Project Request Form
Supervisors: Dr Jianmin Zuo and Professor Martin Rowe
Contact details: x47144
Office Location: S217
School: Cancer Sciences
Study of the dual function immune evasion protein BILF1
Outline of Project:
Despite triggering strong immune responses, Epstein-Barr virus (EBV) has colonized more than 90% of the
adult human population. Successful persistence of EBV depends on the establishment of a balance between
host immune responses and viral immune evasion. A number of immunoevasins targeting the MHC-I and
MHC-II antigen presentation pathways have recently been identified.
One of these immunoevasin genes, BILF1, encodes a viral G Protein-Coupled Receptor (vGPCR) that was
previously shown to physically associate with MHC-I molecules, to target MHC-I for degradation via
lysosomes (1). More recently, Lyngaa et al reported the transforming potential of BILF1 in NIH3T3 cells
in vitro as well as in vivo by using nude mice (2). To understand how BILF1 can be a transforming protein,
we propose to compare the apoptosis and proliferation functions of BILF1 expressing cells line (including B
cell, epithelial cell lines) and control cell lines. To understand how important the BILF1 is in virus-induced
B cell transformation, we propose to carry out the transformation assay by using BILF1 knock out
recombinant EBV compared with wild type EBV and other recombinant EBV.
Techniques to be used in the project:
Production of Recombinant Retrovirus vectors and Recombinant Epstein-Barr virus, Separation of B cells,
Apoptosis Assay, Transformation Assay.
Are these techniques already established in the laboratory where the project would take place?
Yes
Names of other researchers who may also be involved in project (whether supervisory or technical):
Rose Tierney
Key References:
1. Zuo, J., et al., The Epstein-Barr virus G-protein-coupled receptor contributes to immune evasion by
targeting MHC class I molecules for degradation. PLoS Pathog, 2009. 5(1): p. e1000255.
2. Lyngaa, R., et al., Cell transformation mediated by the Epstein-Barr virus G protein-coupled receptor
BILF1 is dependent on constitutive signaling. Oncogene, 2010.
Does this project require the student to hold a personal home office licence?
No
Do you have all the relevant ethical approval for this study?
Yes





