Supplementary Information (doc 2312K)

Supporting Information
Additional Congeners of the Macrolide Neaumycin: Structure Revision and Biological Activity
Matteo Simone
§,
*, Sonia I. Maffioli
§
, Arianna Tocchetti
Biunno
O
, Eleonora Gaspari
§
and Stefano Donadio
§
.
§
, Stefano Tretter
§
, Monica Cattaneo
O
, Ida
§
: NAICONS Srl, Milan, Italy; O : Istitute for Genetic and Biomedical Research (National Council of Research) Milan, Italy.
General Experimental Procedure
Actinoplanes Cultivation and Metabolite Purification
Chemistry
Figure S1.
Antibacterial activity
Cytotoxicity assays
HPLC analysis of an AF1 culture of
Table of Content
Actinoplanes sp. ATCC 33076
Figure S2. HPLC of neaumycin complex
Figure S3. Biomass, neaumycin and ramoplanin A2 production in Actinoplanes sp ATCC 33076.
Figure S4.
Figure S5.
1
Figure S6
LC-MS of amidated compound (
H NMR (CDCl
3
) spectrum of 2
. COSY NMR (CDCl
3
4 ) in positive mode.
) spectrum of 2
Figure S7 . TOCSY NMR (CDCl
3
) spectrum of 2
Figure S8 . HSQC NMR (CDCl
3
) spectrum of 2
Figure S9 . HSQC NMR (CD
3
OD) spectrum of 2
Figure S10 . HMBC NMR (CD
3
OD) spectrum of 2
Figure S11.
13 C NMR (CD
3
OD) spectrum of 2
Figure S12 .
1
H NMR (CDCl
3
) spectrum of 3
Figure S13 . COSY NMR (CDCl
3
) spectrum of 3
Figure S14 . TOCSY NMR (CDCl
3
) spectrum of 3
Figure S15 . HSQC NMR (CDCl
3
) spectrum of 3
Figure S16
1
H NMR (CDCl
3
) spectrum of 1
Figure S17 HSQC NMR (CDCl
3
) spectrum of 1
Table S1. NMR Spectroscopic Data (CDCl
3
) for neaumycin B ( 2 )
Table S2. NMR Spectroscopic Data (CDCl
3
) for neaumycin C ( 3 )
S2
S2
S4
S5
S12
S13
S17
S19
S20
S2
S2
S3
S3
S4
S6
S7
S8
S9
S10
S11
S14
S15
S16
S18
Table S3. NMR Spectroscopic Data (CD
3
OD) for neaumycin B ( 2 )
Table S4. NMR Spectroscopic Data (CDCl
3
) for neaumycin A ( 1 )
S1
General Experimental Procedure
S21
S22
For monitoring metabolites production analytical HPLC was performed on Shimadzu Series 10 spectrophotometer (Kyoto, Japan), equipped with a reverse-phase column, Lichrosorb RP-18, 5μm, 4X250 mm (Merck, Darmstadt, Germany) eluted at 1 ml min-1 at
50°C using a linear gradient of 0.1% (v/v) TFA-acetonitrile from 10 to 90% organic phase in 30 min with detection at 270 nm. For resolution of the neaumycin complex (Figure S2), the above system employed a Mediterranean Sea C18 4.6x150 mm column
(Tecknokroma, Barcelona, Spain) and a linear gradient of 0.1% (v/v) TFA-acetonitrile (from 50 to 90% of organic phase in 15 min and then 90% of organic phase until 25 min). Semi-preparative HPLC employed a Shimadzu Series 6 spectrophotometer, Mediterranean sea C18 10X150 mm column (Tecknokroma, Barcelona, Spain), detection at 270 nm, a linear gradient of eluents A [0.1%
(v/v) TFA] and B (acetonitrile) from 50 to 90% B in 15 min at a flow rate of 1 ml min-1. ESI-MS data were recorded on an Ion
Trap ESQUIRE 3000 Plus spectrometer (Bruker, Karlsruhe, Germany) equipped with an LC Agilent 1100 Diode-Array Detector, using an Ascentis express Supelco RP18, 2,7
µ
(50x4.6 mm) column kept at 40°C, eluting at 1ml min-1 with a 7 min linear gradient from 95:5 phase A [0.05% (v/v) TFA]:phase B [0.05% (v/v) TFA acid in acetonitrile] to 100% phase B.
1
H-,
13
C-, 1- and 2-D NMR spectra (COSY, TOCSY, , HSQC, HMBC) were measured in CDCl
3
or CD
3
OD at 298K using an AMX 400 MHz spectrometer
(Bruker, Karlsruhe, Germany). 1 H and 13 C chemical shifts are reported relative to CDCl
3
(δ 7.26 and 77 ppm) or CD
3
OD (δ 3.34 and 49.86 ppm).
Actinoplanes Cultivation and Metabolite Purification
Actinoplanes sp. ATCC 33076 was propagated on oatmeal agar * and seed cultures were made in AF medium * . For production, a
72-h culture was inoculated (8%) into AF1 medium (a modified AF medium containing 1.0g l -1 CaCO
3
instead of 4.0g l -1 ). Flasks were incubated at 30°C and 200 rpm on a rotary shaker. For 15 l scale fermentation, cultivations in BioFlo® 415 bioreactor (New
Brunswick Scientific) were carried out for 96-120 h at 30°C, with stirring at 500 rpm and 0.4 vvm aeration rate. pH was maintained at 7.3
±
0.2 by adding H
2
SO
4
(5% v/v) , while foam formation was controlled by addition of polypropylene glycol 2000. For dry weight determinations, mycelium from 6 ml culture was separated from broth by filtration on a pre-weighted filter membrane. The mycelium was dried with the filter for 24 h at 37°C. For production analysis, 450 µl were withdrawn and processed by addition of 2 volumes of methanol. Following shaking for 1 h at 40°C and centrifugation (3000 rpm for 10 min), 10 µl of the resulting supernatant was analyzed by HPLC as described above. A culture in AF1 medium (one liter total volume) was filtered through paper filter
(Scienceware, Sigma-Aldrich, St. Louis, MO USA) and the resulting filtrate (0.8 l) was extracted with two volumes of n-butanol.
The separated organic phase was concentrated under vacuum to dryness and purified by multiple preparative TLC chromatography
(Merck, Darmstadt, Germany); compounds were eluted with ethyl acetate as the mobile phase to afford two fractions with RF of 0.5 and 0.3. The first fraction contained 2 (1 mg) while the second one, containing a mixture of 1 and 3 , was further purified by semipreparative HPLC to achieve 3 mg of 1 and 1 mg of 3 .
* Donadio, S; Monciardini, P.; Sosio, M.
Methods. Enzymol.
458, 3-28 (2009).
Chemistry
Preparation of 3-dimethylaminopropylamide of 1 : to a stirred solution of 1 (1 mg) in DMF (1 ml), 3-dimethylamino-propylamine
(0.8 mg 8 eq.), HOBt (1.1 mg 8 eq.) and DCC (1.65 mg, 8 eq.) were added; the reaction was kept under stirring over night at room temperature before LC-MS analysis. A new peak with retention time 4.8 min and m/z 1071.8 [M+H] + was detected corresponding to 4 . When reacted in the same conditions compound 2 resulted unreactive. Hydrolysis: 0.5 mg 1 was dissolved in 500 µl acetonitrile and 500 µl of 0.1N NaOH were added. The reaction was stirred at room temperature for 1h before HPLC-MS analysis.
Antibacterial activity
MICs were determined by the broth microdilution methodology, according to Clinical and Laboratory Standard Institute (CLSI) guidelines ** : bacteria were inoculated at 5x10 5 cfu/ml in Müller Hinton broth containing 20 mg l -1 CaCl
2
and 10mg l -1 MgCl
2
(for
Staphylococcus aureus ) or Todd Hewitt broth (for Streptococcus spp.) in sterile round-bottom, 96-well microtiter plates. Cultures were incubated at 37°C in aerobic condition and read after 20-24 h. Compouds were dissolved in acetonitrile and serial dilutions were made with culture medium immediately before testing. The MIC was defined as the lowest compound concentration causing complete suppression of visible bacterial growth. The effect of neaumycin-ramoplanin combinations was assessed by a two dimensional microdilution assay using 96-well microtiter plates, as above. For each ramoplanin concentration, the lowest neaumycin concentration preventing bacterial growth was recorded.
** Standard National Committee for Clinical Laboratory. ‘’Performance standards for Antimicrobial Susceptibility Testing.’’
Twenty-second Informational Supplement, 2012: CLSI document M100-S22, NCCLS, Wayne, Pennsylvania, USA
S2
Cytotoxicity assays
Human pancreatic carcinoma Panc1 (ATCC CRL-1469) and human embryonic kidney hek-293 (ATCC CRL-1573) were maintained in Dulbecco Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (Euroclone), 2mM glutamine and 100 IU ml -1 of penicillin/streptomycin in a humidified chamber (95% air and 5% CO
2
) at 37°C. Cell viability was determined by using the
Vybrant 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium (MTT) Cell Proliferation Assay Kit (Invitrogen). In brief, cells were seeded at densities of approximately 50-60% in 96-well microplates and treated with Daunorubicin (0.25, 1, 2 and 4 µg ml -1 ), compounds 1 and 2 (1, 10 and 100 μM) for 24 and 48 h. Then, the cells were incubated with 1 mg ml -1 MTT in DMEM supplemented as above for 4 h at 37°C. Formazan release was quantified at 560 nm using a Microplate Reader (Biotek). All absorbance values were corrected by subtraction of the absorbance of non-inoculated wells. The percentage of cell survival was calculated from the absorbance ratio of treated to untreated cells. The percentage survivals are the average of three measurements each from two independent experiments
A2
Ramoplanin congeners
A1 A3
Neaumycin complex
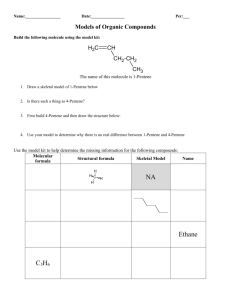
Figure S1: HPLC analysis of an AF1 culture of Actinoplanes sp. ATCC 33076
S3
Figure S2 . HPLC trace of Neaumycin complex before chromatographic purification.
Figure S3 . Biomass (mg), neaumycin and ramoplanin A2 (mg l -1 ) production in Actinoplanes sp ATCC 33076 .The strain was cultured in AF1 medium over 10 days. Each point shown represents the mean of the values obtained from two independent flasks.
S4
Figure S4 . LC-MS (ESI+) analysis of Neaumycin amidation: compound ( 4 ) detected with m/z 1071
M+H
+ and 1094
M+Na
+ ions.
S5
Figure S5 . 1 H NMR (400 MHz, CDCl
3
) spectrum of 2
Compound 2
S6
Figure S6 . COSY NMR (400 MHz, CDCl
3
) spectrum of 2
O
OH
OH
Compound 2
OH
O
O
O
O
O
O O
OH
S7
Figure S7 . TOCSY NMR (400 MHz, CDCl
3
) spectrum of 2.
O
OH
OH
Compound 2
OH
O
O
O
O
O
O O
OH
S8
Figure S8 . HSQC NMR (400 MHz, CDCl
3
) spectrum of 2
O
OH
OH
Compound 2
OH
O
O
O
O
O
O O
OH
S9
Figure S9 . HSQC NMR (400 MHz, CD
3
OD) spectrum of 2
O
OH
OH
Compound 2
OH
O
O
O
O
O
O O
OH
S10
Figure S10 . HMBC NMR (400 MHz, CD
3
OD) spectrum of 2.
O
OH
OH
Compound 2
OH
O
O
O
O
O
O
O
OH
S11
Figure S11 . 13 C NMR (50 MHz, CD
3
OD) spectrum of 2
O
OH
OH
Compound 2
OH
O
O
O
O
O
O O
OH
S12
Figure S12 . 1 H NMR (400 MHz, CDCl
3
) spectrum of 3
Compound 3
S13
Figure S13 . COSY NMR (400 MHz, CDCl
3
) spectrum of 3
Compound 3
S14
Figure S14 . TOCSY NMR (400 MHz, CDCl
3
) spectrum of 3
Compound 3
S15
Figure S15 . HSQC NMR (400 MHz, CDCl
3
) spectrum of 3
Compound 3
S16
Figure S16 . 1 H NMR (CDCl
3
) spectrum of 1
Compound 1
S17
Figure S17 . HSQC NMR (CDCl
3
) spectrum of 1
Compound 1
S18
Table S1. NMR Spectroscopic Data (CDCl
3
) for neaumycin B ( 2 ) position δ
C
, type
44
45
46
47
53
38
39
40
41
42
43
54
55
32
33
34
35
36
37
27
28
29
30
31
21
22
23
24
25
26
15
16
17
18
19
20
9
10
11
12
13
14
4
5
6
1
2
3
7
8
35.5, CH
37.4, CH
2
19.4, CH
2
14.1, CH
3
16.8, CH
3
14.8, CH
3
16.5, CH
3
13.0, CH
3
10.1, CH
3
16.7, CH
3
55.6, CH
3
57.7, CH
3
58.0, CH
3
71.3, CH
35.6, CH
2
,C
37.4, CH
2
19.9, CH
2
30.7, CH
2
72.9, CH
43.1, CH
81.1, CH
60.9, CH
58.6, CH
32.4, CH
2
129.0, CH
133.8, CH
131.4, CH
131.4, CH
133.5, CH
138.7, CH
45.9, CH
76.0, CH
37.5, CH
2
75.7, CH
41.0, CH
166.1, C
121.4, CH
150.2, CH
45.1, CH
75.0, CH
40.0, CH
2
84.2, CH
130.4, CH
136.8, CH
38.3, CH
68.9, CH
39.1, CH
2
69.5, CH
74.0, CH
5.48 dd (15.2, 9)
2,09, m
3.46, m
1.24-2.03, m
3.87 t (10.3)
1,5, m
4.8 dt (11, 4.5)
1.26-2.66, m
1.42-1.65, m
1.39-1.72, m
0.92-1.66, m
3.54, m
1.6, m
3.19, m
2.95, m (7.8)
2.47, m
1.39, m
1.24, m
1.6, m
0.93, m
1.06, m
1.12 d (6.6)
δ
H
, (J in Hz)
5.75, d (15.9)
6.98, dd (16.6,10.4)
2.26, m
3.68 , m
1.43-1.57, m
3.67,m
5.18, dd (15.3, 8.7)
5.50, dd (15.2, 9)
2.2, m
3.89, m
1.44, m
3.48, m
2.92, m
2.17-2.46, m
5.50, m
6.16, m
6.13, m
6.07 , m
5.17 dd (15.3, 8.7)
0.96, m
0.7, d (6.5)
1.02, m
1.06, d (6.3)
3.24 (s)
3.43 (s)
3.57 (s)
S19
Table S2 . NMR Spectroscopic Data (CDCl
3
) for neaumycin C ( 3 )
43, CH
80.4, CH
61.2, CH
58.6, CH
35, CH
36, CH
2
20, CH
2
14, CH
3
17, CH
3
12.4, CH
3
17.1, CH
3 na, CH
137, CH
45, CH
73, CH
37, CH
2
76, CH
41, CH
71, CH
33.7, CH
2 na, C
36, CH
2
20, CH
2
28.7, CH
2
73, CH
13, CH
3
10.4, CH
3
16.7, CH
3
-
57, CH
58, CH
134.5, CH
41.5, CH
70, CH na, CH
2
70, CH
84, CH
32, CH
2 na, CH na, CH na, CH na, CH
δ
C
, type
165, C
122, CH
150, CH
44.8, CH
76, CH
40, CH
2
75, CH na, CH na: not assigned
S20
40
41
42
43
44
34
35
36
37
38
39
45
46
47
53
54
55
28
29
30
31
32
33
20
21
22
23
24
25
26
27
14
15
16
17
18
19
9
10
11
12
13 position
1
2
3
4
5
6
7
8
1.4
0.94
1.12
0.87
1.13
1.63
3.22
2.95
2.5 na
1.3
6.03
5.48
2,2
3.50
1.24-2.01
3.88
1,5
4.85
1.31-2.68
-
1.40-1.68
1.57-1.74
0.97-1.74 na
0.7
0.96
1.04
-
3.43
3.56
5.72
2.25
3.93
1.47
3.76
3.09
2.33-2.56
5.50
6.13 na na
δ
H
-
5.82
6.8
2.39
3.56
1.48-1.57
4.3
5.48
Table S3 . NMR Spectroscopic Data (CD
3
OD) for neaumycin B ( 2 ) position
δ
C
, type
δ
H
43
44
45
46
47
53
54
55
37
38
39
40
41
42
32
33
34
35
36
26
27
28
29
30
31
20
21
22
23
24
25
14
15
16
17
18
19
9
10
11
6
7
8
12
13
1
2
3
4
5 na: not assigned
S21
Table S4 . NMR Spectroscopic Data (CDCl
3
) for neaumycin A ( 1 )
60.6, CH na, CH
37.7, CH
2
19.7, CH
2
14, CH
3
15, CH
3
14.8, CH
3
18, CH
3
14, CH
3
11, CH
3
18, CH
3
56, CH
3
59, CH
3
59.2, CH
3
43, CH
74, CH
35.3, CH
2
100.7, C na, CH
2
19.73, CH
2
28.48, CH
2 na, CH
44.7, CH
82.7, CH
62.5, CH
86.4, CH na, CH
2
131, CH
134.6, CH na, CH na, CH
133, CH
138, CH na, CH na, CH n.a., CH
2
75.4, CH
168, C
122, CH
152, CH
44.9, CH
73.5, CH
40.6, CH
2
83, CH
130.7, CH
140, CH na, CH
72.3, CH na, CH
2
71.1, CH
1.02
1.16
0.76
0.99
1.05
3.28
3.4
3.52
2.54 na na
1.43
0.95
1.11
1,47
4.75
1.27-2.78
-
1.5-1.75
1.44-1.77
1.07-1.78 na
1.7
3.27
3.01
6.08
5.46 na
3.50 na
3.78
3.04
2.34
5.64
6.21 na na
-
5.86
6.88
2.33 na
1.48-1.57
3.8
5.24
5.77 na
3.83 na
3.73
S22
43.6, CH
80, CH
63.3, CH
59.6, CH
34.6, CH
35.6, CH
2
20.7, CH
2
13.8, CH
3
17.2, CH
3
10.4, CH
3
17.1, CH
3
13.3, CH
3
131.4, CH
138, CH
44.7, CH
70, CH
37.4, CH
2
77, CH
28, CH
71, CH
34, CH
2
99.1, C
36, CH
2
20.4, CH
2
29, CH
2
72.7, CH
10.1, CH
3
18.6, CH
3
55.6, CH
3
57.7, CH
3
58, CH
3
138.4, CH
39.8, CH
67, CH
37.6, CH
2
74, CH
80, CH
34, CH
2
127.5, CH
134.7, CH
131.1, CH
132.2, CH
δ
C
, type
166.5, C
121, CH
152, CH
43.9, CH
75.6, CH
40.1, CH
2
85, CH
128, CH
1.58
3.34
3.18
2.57
1.5
1.25-1.41
1.44
0.94
1.13
1.04
1.26
0.82
6.03
5.3
2.15
3.31
1.28-2.13
3.93
1.5
4.88
1.31-2.54
-
1.4-1.68
1.53-1.75
0.99-1.71
3.49
0.98
1.04
3.24
3.43
3.57
5.76
2.17
3.34
1.71-1.91
4.95
3.38
2.78
5.29
5.74
6.06
5.9
δ
H
-
5.81
6.78
2.31
3.6
1.48-1.58
3.75
5.26
40
41
42
43
44
45
34
35
36
37
38
39
46
47
53
54
55
26
27
28
29
30
31
32
33
20
21
22
23
24
25
15
16
17
18
19
9
10
11
12
13
14 position
1
2
6
7
8
3
4
5