Instructor`s Manual to accompany Principles of Life
advertisement

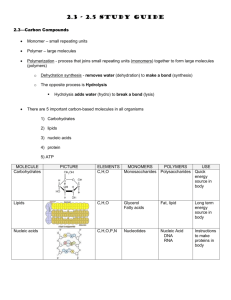
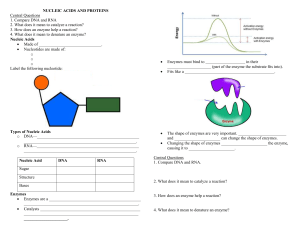
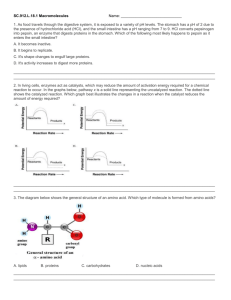
Principles of Life Hillis • Sadava • Heller • Price Instructor’s Manual Chapter 3: Nucleic Acids, Proteins, and Enzymes OVERVIEW Chapter 3 provides detailed descriptions of the structure of nucleic acids and proteins. How DNA and RNA are constructed from nucleotides, including base pairing, is described. DNA and RNA similarities and differences are also included. Amino acids and the complex structure of proteins, including primary, secondary, tertiary, and quaternary structure, are described. Enzyme function is discussed, including how structure defines the function of enzymes. How enzymes catalyze reactions and what determines reaction rates is included, along with regulation of enzymes and metabolic pathways. Factors that affect protein denaturing are also discussed. KEY CONCEPTS/CHAPTER OUTLINE 3.1 Nucleic Acids Are Informational Macromolecules • Nucleotides are the building blocks of nucleic acids • Base pairing occurs in both DNA and RNA • DNA carries information and is expressed through RNA Nucleotides consist of a nitrogen-containing base, a pentose sugar, and phosphate groups. They are linked by phosphodiester linkages in condensation reactions to form RNA and DNA. Complementary base pairing by hydrogen bonding is the key to structure and function of these molecules. RNA can be a single strand, or folded into a 3-D structure. RNA is important in the transcription and translation of DNA information, and also helps regulate gene expression. DNA is usually a double strand that is twisted into a double helix. DNA can self-replicate, and carries information in the sequence of bases for building proteins. Biologists can compare the base sequences of organisms to determine evolutionary relationships. 3.2 Proteins Are Polymers with Important Structural and Metabolic Roles • Amino acids are the building blocks of proteins • Amino acids are bonded to one another by peptide linkages • Higher-level protein structure is determined by primary structure • Environmental conditions affect protein structure Proteins are polymers of about 20 different amino acids, and have many roles in organisms, including structure, storage, defense, transport, signaling, and as enzymes. Each amino acid has a side chain or R group with different characteristics. Peptide linkages link amino acids to form the primary structure of a protein—the sequence of amino acids determines the structure and function of the protein. Secondary and tertiary structure result from protein folding, and interaction of the amino acid side chains to form 3-D structures. Quaternary structure results © 2012 Sinauer Associates, Inc. 1 from binding of folded subunits (polypeptides) into complex proteins. Loss of higher level structure (denaturing) can result from high temperatures or change in pH. 3.3 Some Proteins Act as Enzymes to Speed up Biochemical Reactions • To speed up a reaction, an energy barrier must be overcome • Enzymes bind specific reactants at their active sites Enzymes act as catalysts to reduce activation energy and increase the rate of metabolic reactions. The enzyme is not changed during the reaction. Enzymes are highly specific and bind only one or a few reactants. Enzymes work by bringing reactants close together, adding chemical groups, or inducing strain in the reactant molecules. Some enzymes require cofactors, coenzymes, or prosthetic groups to function. The rate of enzyme-catalyzed reactions levels off when all enzyme active sites are full or saturated. 3.4 Regulation of Metabolism Occurs by Regulation of Enzymes • Enzymes can be regulated by inhibitors • An allosteric enzyme is regulated via changes in its shape • Some metabolic pathways are usually controlled by feedback inhibition • Enzymes are affected by their environments In metabolic pathways, the product of one reaction is a substrate for the next. Each step is catalyzed by a different enzyme. Maintenance of homeostasis is accomplished by regulating metabolic pathways, by regulating enzymes. Inhibitors bind to enzymes and prevent substrate from binding. In allosteric regulation, the shape of the active site changes when a molecule binds to the enzyme. The change in shape either allows substrate to bind, or prevents it. Some pathways are regulated by feedback inhibition— in which the end-product binds to the first enzyme in the pathway. Temperature and pH can cause denaturing by breaking noncovalent interactions, and by changing hydrophilic and hydrophobic interactions. Some organisms produce isozymes—different forms of enzymes that function at different optimal conditions— allowing organisms to adapt to environmental changes. LECTURE OUTLINE Chapter 3 Opening Question How does an understanding of proteins and enzymes help to explain how aspirin works? Concept 3.1 Nucleic Acids Are Informational Macromolecules Nucleic acids are polymers specialized for storage, transmission, and use of genetic information. DNA = deoxyribonucleic acid RNA = ribonucleic acid Monomers: Nucleotides Nucleotide: Pentose sugar + N-containing base + phosphate group Nucleosides: Pentose sugar + N-containing base FIGURE 3.1 Nucleotides Have Three Components Bases: © 2012 Sinauer Associates, Inc. 2 Pyrimidines—single rings Purines—double rings Sugars: DNA contains deoxyribose RNA contains ribose FIGURE 3.1 Nucleotides Have Three Components Nucleotides bond in condensation reactions to form phosphodiester linkages. Nucleic acids grow in the 5′ to 3′ direction. FIGURE 3.2 Linking Nucleotides Together Oligonucleotides have about 20 monomers, and include small RNA molecules important for DNA replication and gene expression. DNA and RNA are polynucleotides, the longest polymers in the living world. TABLE 3.1 Distinguishing RNA from DNA Complementary base pairing: adenine and thymine always pair (A-T) cytosine and guanine always pair (C-G) IN-TEXT ART, p. 36 Base pairs are linked by hydrogen bonds. There are so many hydrogen bonds in DNA and RNA that they form a fairly strong attraction, but not as strong as covalent bonds. Thus, base pairs can be separated with only a small amount of energy. (LINK Hydrogen bonds play an essential role in the chemistry of life; see Concept 2.2) RNA is usually single-stranded, but may be folded into 3-D structures, by hydrogen bonding. Folding occurs by complementary base pairing, so structure is determined by the order of bases. FIGURE 3.3 RNA DNA: Two polynucleotide strands form a “ladder” that twists into a double helix. Sugar-phosphate groups form the sides of the ladder, the hydrogen-bonded bases form the rungs. (VIDEO 3.1 Deoxyribonucleic acid: a three-dimensional model) (VIDEO 3.2 Cell visualization: the structure of DNA) FIGURE 3.4 DNA DNA is an informational molecule: genetic information is in the sequence of base pairs. DNA has two functions: Replication © 2012 Sinauer Associates, Inc. 3 Gene expression—base sequences are copied to RNA, and specify amino acids sequences in proteins. IN-TEXT ART, p. 37 DNA replication and transcription depend on the base pairing: 5′-TCAGCA-3′ 3′-AGTCGT-5′ 3′-AGTCGT-5′ transcribes to RNA with the sequence 5′-UCAGCA-3′. Genome: Complete set of DNA in a living organism Genes: DNA sequences that encode specific proteins and are transcribed into RNA Not all genes are transcribed in all cells of an organism. FIGURE 3.5 DNA Replication and Transcription DNA base sequences reveal evolutionary relationships. Closely related living species should have more similar base sequences than species that are more distantly related. Scientists are now able to determine and compare entire genomes of organisms to study evolutionary relationships. (ANIMATED TUTORIAL 3.1 Macromolecules: Nucleic Acids and Proteins) (LINK The use of DNA sequences to reconstruct the evolutionary history of life is described in Chapter 16) Concept 3.2 Proteins Are Polymers with Important Structural and Metabolic Roles Major functions of proteins: • Enzymes—catalytic proteins • Defensive proteins (e.g., antibodies) • Hormonal and regulatory proteins—control physiological processes • Receptor proteins—receive and respond to molecular signals • Storage proteins store amino acids • Structural proteins—physical stability and movement • Transport proteins carry substances (e.g., hemoglobin) • Genetic regulatory proteins regulate when, how, and to what extent a gene is expressed Protein monomers are amino acids. Amino and carboxylic acid functional groups allow them to act as both acid and base. The R group differs in each amino acid. Only 20 amino acids occur extensively in the proteins of all organisms. They are grouped according to properties conferred by the R groups. TABLE 3.2 The Twenty Amino Acids in Proteins Cysteine side chains can form covalent bonds: a disulfide bridge, or disulfide bond. © 2012 Sinauer Associates, Inc. 4 Oligopeptides or peptides: Short polymers of 20 or fewer amino acids—some hormones and signaling molecules Polypeptides or proteins range in size from insulin, which has 51 amino acids, to huge molecules such as the muscle protein titin, with 34,350 amino acids. Amino acids are linked in condensation reactions to form peptide linkages or bonds. Polymerization takes place in the amino to carboxyl direction. FIGURE 3.6 Formation of a Peptide Linkage Primary structure of a protein: The sequence of amino acids Secondary structure: Regular, repeated spatial patterns in different regions, resulting from hydrogen bonding • α (alpha) helix—right-handed coil • β (beta) pleated sheet—two or more polypeptide chains are extended and aligned FIGURE 3.7 The Four Levels of Protein Structure Tertiary structure: Polypeptide chain is bent and folded; results in the definitive 3-D shape The outer surfaces present functional groups that can interact with other molecules. (VIDEO 3.3 Lysozyme: A three-dimensional model) Interactions between R groups determine tertiary structure. • Disulfide bridges hold a folded polypeptide together • Hydrogen bonds stabilize folds • Hydrophobic side chains can aggregate • van der Waals interactions between hydrophobic side chains • Ionic interactions form salt bridges FIGURE 3.8 Noncovalent Interactions between Proteins and Other Molecules FIGURE 3.9 The Structure of a Protein Secondary and tertiary protein structure derive from primary structure. Denaturing: Heat or chemicals are used to disrupt weaker interactions in a protein, destroying secondary and tertiary structure. The protein can return to normal when cooled—all the information needed to specify the unique shape is contained in the primary structure. FIGURE 3.10 Primary Structure Specifies Tertiary Structure Quaternary structure: Two or more polypeptide chains (subunits) bind together by hydrophobic and ionic interactions, and hydrogen bonds. These weak interactions allow small changes that aid in the protein’s function. © 2012 Sinauer Associates, Inc. 5 IN-TEXT ART, p. 44 (VIDEO 3.4 Hemoglobin: A three-dimensional model) FIGURE 3.7 The Four Levels of Protein Structure Factors that can disrupt the interactions that determine protein structure (denaturing): • Temperature • Concentration of H+ • High concentrations of polar substances • Nonpolar substances (APPLY THE CONCEPT Proteins are polymers with important structural and metabolic roles) Concept 3.3 Some Proteins Act as Enzymes to Speed up Biochemical Reactions Living systems depend on reactions that occur spontaneously, but at very slow rates. Catalysts are substances that speed up reactions without being permanently altered. No catalyst makes a reaction occur that cannot otherwise occur. Most biological catalysts are proteins (enzymes); a few are RNA molecules (ribozymes). In some exergonic reactions there is an energy barrier between reactants and products. An input of energy (the activation energy or Ea) will put reactants into a transition state. FIGURE 3.11 Activation Energy Initiates Reactions Enzymes lower the activation energy—they allow reactants to come together and react more easily. Example: A molecule of sucrose in solution may hydrolyze in about 15 days; with sucrase present, the same reaction occurs in 1 second! FIGURE 3.12 Enzymes Lower the Energy Barrier Enzymes are highly specific—each one catalyzes only one chemical reaction. Reactants are substrates: they bind to a specific site on the enzyme—the active site. Specificity results from the exact 3-D shape and chemical properties of the active site. FIGURE 3.13 Enzyme Action The enzyme–substrate complex (ES) is held together by hydrogen bonding, electrical attraction, or temporary covalent bonding. E S ES E P The enzyme is not changed at the end of the reaction. Enzymes may use one or more mechanisms to catalyze a reaction: • Inducing strain: Bonds in the substrate are stretched, putting it in an unstable transition state. IN-TEXT ART, p. 47 • Substrate orientation: Substrates are brought together so that bonds can form. © 2012 Sinauer Associates, Inc. 6 • Adding chemical groups: R groups may be directly involved in the reaction. Binding of substrate to enzyme is like a baseball in a catcher’s mitt. The enzyme changes shape to make the binding tight—“induced fit”. FIGURE 3.14 Some Enzymes Change Shape When Substrate Binds to Them Some enzymes require ions or other molecules in order to function: • Cofactors—inorganic ions • Coenzymes add or remove chemical groups from the substrate. They can participate in many different reactions. • Prosthetic groups (non–amino acid groups) permanently bound to their enzymes. (VIDEO 3.5 Glyceraldehyde 3-phosphate dehydrogenase and coenzyme NAD: A threedimensional model) TABLE 3.3 Some Examples of Nonprotein “Partners” of Enzymes Rates of catalyzed reactions: There is usually less enzyme than substrate present, so reaction rate levels off when the enzyme becomes saturated. Saturated—all enzyme molecules are bound to substrate molecules. FIGURE 3.15 Catalyzed Reactions Reach a Maximum Rate Maximum rate is used to calculate enzyme efficiency—molecules of substrate converted to product per unit time (turnover). It ranges from 1 to 40 million molecules per second! Concept 3.4 Regulation of Metabolism Occurs by Regulation of Enzymes Enzyme-catalyzed reactions are part of metabolic pathways—the product of one reaction is a substrate for the next. Homeostasis—the maintenance of stable internal conditions Cells can regulate metabolism by controlling the amount of an enzyme. Cells often have the ability to turn synthesis of enzymes off or on. (LINK The regulation of enzyme synthesis is described in Chapter 11) Chemical inhibitors can bind to enzymes and slow reaction rates. Natural inhibitors regulate metabolism; artificial inhibitors are used to treat diseases, kill pests, and study enzyme function. Irreversible inhibition—inhibitor covalently binds to a side chain in the active site. The enzyme is permanently inactivated. FIGURE 3.16 Irreversible Inhibition © 2012 Sinauer Associates, Inc. 7 Reversible inhibition (more common in cells): A competitive inhibitor competes with natural substrate for active site. A noncompetitive inhibitor binds at a site distinct from the active site—this causes change in enzyme shape and function. (ANIMATED TUTORIAL 3.2 Enzyme Catalysis) FIGURE 3.17 Reversible Inhibition Allosteric regulation: Non-substrate molecule binds a site other than the active site (the allosteric site) The enzyme changes shape, which alters the chemical attraction (affinity) of the active site for the substrate. Allosteric regulation can activate or inactivate enzymes. (ANIMATED TUTORIAL 3.3 Allosteric Regulation of Enzymes) FIGURE 3.18 Allosteric Regulation of Enzyme Activity Protein kinases are enzymes that regulate responses to the environment by organisms. They are subject to allosteric regulation. The active form regulates the activity of other enzymes, by phosphorylating allosteric or active sites on the other enzymes. (LINK The role of protein kinases in intracellular signaling pathways is described in Chapter 5) Metabolic pathways: The first reaction is the commitment step—other reactions then happen in sequence. Feedback inhibition (end-product inhibition): The final product acts as a noncompetitive inhibitor of the first enzyme, which shuts down the pathway. FIGURE 3.19 Feedback Inhibition of Metabolic Pathways pH affects enzyme activity: Acidic side chains generate H+ and become anions. Basic side chains attract H+ and become cations. Example: glutamic acid—COOH glutamic acid—COO– + H+ The law of mass action: The higher the H+ concentration, the more reaction is driven to the left to the less hydrophillic form—this can affect enzyme shape and function. Protein tertiary structure (and thus function) is very sensitive to the concentration of H+ (pH) in the environment. All enzymes have an optimal pH for activity. FIGURE 3.20 A Enzyme Activity Is Affected by the Environment © 2012 Sinauer Associates, Inc. 8 Temperature affects enzyme activity: Warming increases rates of chemical reactions, but if temperature is too high, non-covalent bonds can break and inactivate enzymes. All enzymes have an optimal temperature for activity. FIGURE 3.20 Enzyme Activity Is Affected by the Environment Isozymes catalyze the same reaction but have different composition and physical properties. Isozymes may have different optimal temperatures or pH, allowing an organism to adapt to changes in its environment. (APPLY THE CONCEPT Regulation of metabolism occurs by regulation of enzymes) Answer to Opening Question Aspirin binds to and inhibits the enzyme cyclooxygenase. Cyclooxygenase catalyzes the commitment step for metabolic pathways that produce: Prostaglandins—involved in inflammation and pain Thromboxanes—stimulate blood clotting and constriction of blood vessels FIGURE 3.21 Aspirin: An Enzyme Inhibitor Aspirin binds at the active site of cyclooxygenase and transfers an acetyl group to a serine residue. Serine becomes more hydrophobic, which changes the shape of the active site and makes it inaccessible to the substrate. FIGURE 3.22 Inhibition by Covalent Modification KEY TERMS activation energy (Ea) active site adenine (A) allosteric regulation α (alpha) helix amino acids (beta) pleated sheet base catalysts competitive inhibitor complementary base pairing cytosine (C) denatured deoxyribose disulfide bridge DNA enzyme–substrate complex (ES) © 2012 Sinauer Associates, Inc. 9 feedback inhibition genes genome guanine (G) noncompetitive inhibitor nucleic acids nucleotide peptide linkage phosphodiester linkage primary structure purine pyrimidine quaternary structure R group ribose RNA secondary structure substrates tertiary structure thymine (T) transition state uracil (U) © 2012 Sinauer Associates, Inc. 10