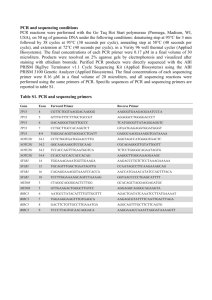
Supplemental Text (online only) DNA and RNA isolation from tumor
advertisement

Supplemental Text (online only) DNA and RNA isolation from tumor specimens Core biopsy specimens were stored in RNALater. Tissues were transferred to a tube containing Qiagen RLT buffer and were homogenized using an electronic homogenizer. Tubes containing the homogenized tissue were centrifuged for 3 minutes at 11,500 rpm to pellet debris. The lysate was subjected to RNA and DNA extraction using the Simultaneous RNA and DNA Extraction (RNALater 76154 RNeasy 74103, 74104, 74106 QiAMP DNA Mini Kit 51304, 79654 DNase Set 79254). RNA and DNA samples were stored at –80°C prior to use. Roche’s AmpliChip TP53 Assay The AmpliChip TP53 Assay is under development at Roche Molecular Systems, Inc. The AmpliChip TP53 test reagents were used to amplify products encompassing the coding regions of the TP53 gene in two reactions (A and B) for all samples, including a reference wild-type DNA. The pooled products generated from the A and B reactions were cleaved by a mix containing DNase I. Fragmentation was performed by recombinant DNase I to generate fragments of an average size of 50 to 100 nucleotides. The fragmented DNA amplicons were subsequently labeled with biotin at their 3’ termini by the action of terminal transferase using AmpliChip TdT labeling reagent as substrate. The biotin-labeled TP53 target DNA fragments were added to the hybridization buffer containing the AmpliChip oligonucleotide solution, which functions as a hybridization control. The mixture was hybridized to the oligonucleotides located on the AmpliChip TP53 Microarray using the Affymetrix GeneChip Fluidics Station 450Dx and an AmpliChip TP53-specific protocol. The hybridized AmpliChip TP53 Microarray was washed and stained with a streptavidin-conjugated fluorescent dye (phycoerythrin). After staining, the AmpliChip TP53 Microarray was scanned by an Affymetrix GeneChip Scanner 3000Dx using a laser that excites the fluorescent label bound to the hybridized TP53 target DNA fragments. The amount of emitted light is proportional to bound target DNA at each location on the probe microarray. Chip design The AmpliChip TP53 microarray was designed by Roche Molecular Systems, Inc., and manufactured by Affymetrix. The microarray consists of a square grid of 228,484 probes. Each probe contains multiple copies of a specific oligonucleotide sequence. A single probe set for an interrogating base position includes five probes, one probe to hybridize to the wild type, three probes to detect three possible single base pair mutations, and one probe to detect single deletion. There are at least 24 probe sets for each nucleotide position, including both sense and antisense probe sequences. A total of 1,240 nucleotide positions of coding regions of exons 2 to 11 and the splice sites are interrogated by AmpliChip TP53. Data analysis of microarray signals The TP53 mutation status was determined by a TP53 Mutation Detection Algorithm developed by Roche Molecular Systems, Inc., which is designed to detect single base pair substitutions and single base pair deletions of a sample in a background of wild-type TP53 DNA probe intensities. The Mutation Detection Algorithm first reads the probe intensities generated by the GeneChip Operating Software provided by Affymetrix. The algorithm first performs the chip quality test using control probes as well as the TP53 probes. If the chip fails the test, no further computation is made. If the chip passes the chip quality test, the algorithm performs quality tests for each PCR product. If a PCR product fails the quality test, the PCR product failure is reported, and no further analysis is made for the PCR product. If a PCR product passes the test, the probe intensities are normalized using quantile normalization to correct chip-to-chip intensity variability. The algorithm makes a tentative call for each base position. Possible base calls for each nucleotide position are wild type, single base substitution, or single base deletion. In the case of a single base substitution, the algorithm identifies the mutated base (eg, G to A). After the tentative calls are made, the reliability of each base call is re-examined using the sequence and key signals from neighbor sites. The algorithm makes adjustments depending on the reliability. Supplemental Fig. 1 Mutation by type. Online only Supplemental Fig. 2 Mutation frequency by exon. Online only