CURRICULUM VITAE
advertisement

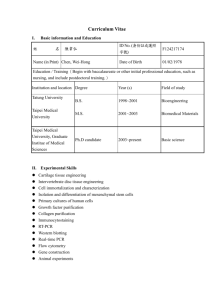
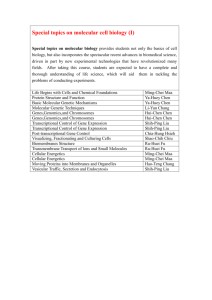
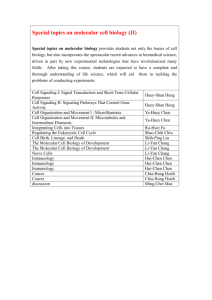
CURRICULUM VITAE Name: Address: Jigang Li 165 Prospect St., OML 352 Department of Molecular, Cellular and Developmental Biology Yale University New Haven, CT 06520 USA Telephone: +1(203)432-8909 (Lab) +1(203)376-3469 (Cell Phone) Email: jigang.li@yale.edu ♦EDUCATION♦ Sep 2001 – Jun 2006 Sep 1997 – Jun 2001 Ph.D., College of Life Sciences, Peking University, Beijing, China B.E., College of Biological Engineering, East China University of Science and Technology, Shanghai, China ♦RESEARCH EXPERIENCE♦ Jan 2011 – present Associate Research Scientist, Department of Molecular, Cellular, and Developmental Biology, Yale University Jan 2007 – Jan 2011 Postdoctoral Associate, Department of Molecular, Cellular and Developmental Biology, Yale University Jul 2006 – Jan 2007 Postdoctoral Research Associate, Peking-Yale Joint Center for Plant Molecular Genetics and Agro-Biotechnology, and National Laboratory of Protein Engineering and Plant Genetic Engineering, Peking University, Beijing, China Aug 2001 – Jul 2006 Ph.D. student, Peking-Yale Joint Center for Plant Molecular Genetics and Agro-Biotechnology, and National Laboratory of Protein Engineering and Plant Genetic Engineering, Peking University, Beijing, China ♦SCIENTIFIC PUBLICATIONS♦ 1) He G*, Chen B*, Wang X*, Li X*, Li J*, He H, Yang M, Lu L, Qi Y, Wang X, Deng XW (2012). Conservation and divergence of transcriptomic and epigenomic variations in maize hybrids. Submitted to Plant Cell. (* equal contribution) 2) Huang X, Ouyang X, Yang P, Lau OS, Li G, Li J, Deng XW (2012). Arabidopsis FHY3 and HY5 positively mediate the COP1 transcription induction in response to photomorphogenic UV-B light. Plant Cell, in press. 3) Zhong S, Shi H, Xue C, Wang L, Xi Y, Li J, Quail PH, Deng XW, Guo H (2012). A molecular framework of light-controlled phytohormone action in Arabidopsis. Current Biology 22:1530-1535. This paper was featured in: Lorrain S, Fankhauser C. Plant development: should I stop or should I grow? Current Biology 22: R645-647. 4) Chen F, Shi X, Chen L, Dai M, Zhou Z, Shen Y, Li J, Li G, Wei N, Deng XW (2012). Phosphorylation of FAR-RED ELONGATED HYPOCOTYL1 is a key mechanism defining signaling dynamics of phytochrome A under red and far-red light. Plant Cell 24:1907-1920. 5) Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li J, Deng XW, Lee CM, Thomashow MF, Yang Y, He Z, He SY (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 109:E1192-1200. 6) Li J, Terzaghi W, Deng XW (2012). Genomic basis for light control of plant development. Protein Cell 3:106-116. 7) Shen H*, He H*, Li J*, Chen W, Wang X, Guo L, Peng Z, He G, Zhong S, Qi Y, Terzaghi W, Deng XW (2012). Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell, 24:875-892. (* equal contribution) This paper was featured in: Hofmann NR. A global view of hybrid vigor: DNA methylation, small RNAs, and gene expression. Plant Cell 24:841. “Author Profile” could be found in: http://www.plantcell.org/content/24/3/875/suppl/DC2 8) Li J, Li G, Wang H, and Deng XW (2011). Phytochrome signaling mechanisms. The Arabidopsis Book 9:e148. doi: 10.1199/tab.0148. 9) Ouyang X*, Li J*, Li G*, Li B*, Chen B, Shen H, Huang X, Mo X, Wan X, Lin R, Li S, Wang H, and Deng XW (2011). Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell 23:2514-2535. (* equal contribution) 10) Li G, Siddiqui H, Teng Y, Lin R, Wan X, Li J, Lau OS, Ouyang X, Dai M, Wan J, Devlin PF, Deng XW and Wang H (2011). Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nature Cell Biology 13:616-622. 11) Li J, Li G, Gao S, Martinez C, He G, Zhou Z, Huang X, Lee JH, Zhang H, Shen Y, Wang H and Deng XW (2010). Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22:3634-3649. 12) Lee JH, Yoon HJ, Terzaghi W, Martinez C, Dai M, Li J, Byun MO and Deng XW (2010). DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell 22:1716-1732. 13) Chen H, Huang X, Gusmaroli G, Terzaghi W, Lau OS, Yanagawa Y, Zhang Y, Li J, Lee JH, Zhu D and Deng XW (2010). Arabidopsis CULLIN4-Damaged DNA Binding Protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 22:108-123. 14) Shen Y, Zhou Z, Feng S, Li J, Tan-Wilson A, Qu LJ, Wang H and Deng XW (2009). Phytochrome A mediates rapid red light–induced phosphorylation of Arabidopsis FAR-RED ELONGATED HYPOCOTYL1 in a low fluence response. Plant Cell 21:494-506. 15) Saijo Y, Zhu D, Li J, Rubio V, Zhou Z, Shen Y, Hoecker U, Wang H and Deng XW (2008). Arabidopsis COP1/SPA1 complex and FHY1/FHY3 associate with distinct phosphorylated forms of phytochrome A in balancing light signaling. Molecular Cell 31:607-613. 16) Yang X, Li J, Pei M, Gu H, Chen Z and Qu LJ (2007). Over-expression of a flower-specific transcription factor gene AtMYB24 causes aberrant anther development. Plant Cell Reports 26:219-228. 17) Li J, Li X, Guo L, Lu F, Feng X, He K, Wei L, Chen Z, Qu LJ and Gu H (2006). A subgroup of MYB transcription factor genes undergoes highly conserved alternative splicing in Arabidopsis and rice. Journal of Experimental Botany 57:1263-1273. 18) Li J, Yang X, Wang Y, Li X, Gao Z, Pei M, Chen Z, Qu LJ and Gu H (2006). Two groups of MYB transcription factors share a motif which enhances trans-activation activity. Biochemical and Biophysical Research Communications 341:1155-1163. 19) Chen Y, Yang X, He K, Liu M, Li J, Gao Z, Lin Z, Zhang Y, Wang X, Qiu X, Shen Y, Zhang L, Deng X, Luo J, Deng XW, Chen Z, Gu H and Qu LJ (2006). The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Molecular Biology 60:107-124. 20) Qin G, Kang D, Dong Y, Shen Y, Zhang L, Deng X, Zhang Y, Li S, Chen N, Niu W, Chen C, Liu P, Chen H, Li J, Ren Y, Gu H, Deng XW, Qu LJ and Chen Z (2003) Obtaining and analysis of flanking sequences from T-DNA transformants of Arabidopsis. Plant Science 165:941-949. ♦REFERENCES♦ Xing Wang Deng, Ph.D. The Daniel C. Eaton Professor of Plant Biology Department of Molecular, Cell, and Development Biology Yale University, P.O. Box 208104 165 Prospect Street, OML 352A New Haven, CT 06520-8104 Phone: 1-203-432-8908; Fax: 1-203-432-5726 Email: xingwang.deng@yale.edu Hongya Gu, Ph.D. Professor and Vice Dean College of Life Sciences Peking University Beijing 100871 China Phone: 86-10-62751847; Fax: 86-10-62751841 Email: guhy@pku.edu.cn Li-Jia Qu, Ph.D. Professor and Associate Director Peking-Yale Joint Center for Plant Molecular Genetics and Agrobiotechnology Peking University Beijing 100871 China Phone: 86-10-62753018; Fax: 86-10-62753339 Email: qulj@pku.edu.cn