A Computer Model of Temperature Selection PCR
advertisement

A Computer Model of Temperature Selection PCR
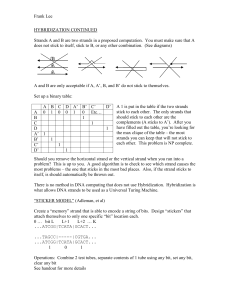
A test tube contains DNA strands of the design shown in Fig. 1. Different strands
may have distinct sequences. Each distinct sequence may be present in millions (or more)
copies. Let us primarily focus on what we will refer to as the “upper” strands. These are
the strands shown as the arrows pointing to the right in Fig. 1. They are characterized by
beginning with the primer sequence S1. Typically, there are greater numbers of some
upper strands than others.
A key design feature is that all the strands have universal, unchanging end
sequences, labeled S1 and S2. These primer sites are carefully designed to dependably
bind to their complementary sequences only when they are in correct alignment.
Naturally, one benefit of universal ends is that an entire library can be readily amplified
using conventional PCR. Optionally, this could be followed by cleavage to remove a
primer site (or both primer sites).
Fig. 1 In this figure, primers are shown as short arrows. Primers are available in surplus.
If and when long (partially or fully)bound strands are separated, primers can bind to their
ends. At that time, polymerase can extend such a primer to a full-length strand that is the
exact complement of the long strand. Appropriate thermal cycling can repeatedly double
the number of copies of all the long strands. This is the usual polymerase chain reaction,
PCR. In the protocol outlined in the figure, temperatures are limited so that only very
mismatched DNA sequences cam be repeatedly copied. Figure \copyright 2001, David
Harlan Wood, by permission.
We may assume that for each upper strand in the test tube, there is a
corresponding lower strand. This is because one starts by synthesizing the upper strands.
Then one adds polymerase and many copies of primer S2’. Polymerase extends a
hybridized primer into complementary lower strand using the upper strand for a template.
From that time on, we make sure that both primers are always present. This means that
whenever an upper strand is being used as a template, its complementary lower strand is
also being used as a template because both primers are always present. As a result, every
upper strand continues to have a corresponding complementary lower strand.
At high temperatures, all the DNA strands melt apart. It is well known that if we
cool the test tube slowly, each upper strand will seek out and bind strongly to its
corresponding complementary lower strand. In temperature selection PCR, the test tube is
cooled quickly, so that strands do not have time often have time to find their exact
complements. Instead, upper strands must quickly bind with lower strands. We call this
"random pairing," or "quenching."
Random bindings vary in strength. Our main objective is to end up with a
collection of DNA strands that bind only weakly to each other. Our protocol is designed
to replicate only the pairs that bind weakly. Strongly bound strands remain in the test
tube, but they do not replicate.
Let us now categorize all possible random pairings. Each group of distinct upper
strands is distributed among the groups of distinct lower strands in proportion to the sizes
of these groups. The distribution of upper strands is modeled as follows. The probability
of upper strand i pairing with lower strand j is
probability i,j = ith proportion of upper strands \times jth proportion of lower
strands .
Since there is one upper strand in each pair, the expected number of i,j pairs is
probability i,j \times total number of upper strands.
For example, suppose there are millions of strands, but each one is one of four
distinct types. Further suppose these four types of distinct strands are present in
proportion to [4, 3, 2, 1]. Since we specify proportions, the units of counting are arbitrary.
Dividing by the sum of 4 + 3 + 2 + 1, we obtain [.4, .3, .2, .1], or 40%, 30%, 20%, and
10%. Proportions of pairs are given by
U
P
P
E
R
0.40
0.30
0.20
0.10
0.40
0.16
0.12
0.08
0.04
0.30
0.12
0.09
0.06
0.03
0.20
0.08
0.06
0.04
0.02
0.10
0.04
0.03
0.02
0.01
L
O
W
E
R
One can readily check that the total of the columns is [.4, .3, .2, .1], and the total of all
entries of the matrix is 1. This verifies that all upper strands are accounted for in the
above matrix.
Successive generations of such paring is illustrated for an example in the authors’
videoclip http://www.thegoldensun.com/dna/dna1.avi
Our intention is that some parings will replicate and some will not. We can
calculate in advance how strongly each pair will bind---and set a threshold for replication.
Several methods of varying sophistication are available for estimating the likelihood of
binding. In any case, we form a selection matrix $S$ with its i,j element zero if strands i
and j are unlikely to bind, and nonzero otherwise. The selection matrix $S$ is multiplied
elementwise with the matrix describing the categories of pairing like the one presented
above. The remaining nonzero pairs of this product are the only ones that replicate
themselves.
A Simplified Model of Hybridization
The model of temperature selection PCR given above can use any selection
matrix $S.$ The i,j element of the Selection matrix $S$ is zero if the ith lower strand
binds to the jth upper strand well enough to prevent their melting apart for replication.
Nonzero elements of $S$ permit replication.
To obtain a qualitative understanding of the temperature selection PCR protocol,
we use a simplified model of hybridization for forming a selection matrix. We simply
count what fractional part of the upper and lower strands are complementary. This
selection matrix estimates the likelihood of replication by simply counting the
mismatched bases in each paring of an upper and a lower strand. This method is best
suited to DNA sequences consisting of only A and T. Since C and G have different
binding power than A with T, merely counting mismatches is cruder for sequences using
more than these two bases.
Randomly chosen upper and lower strands will tend to hybridize because each
primer sequence encounters its exact complement. We assume that no upper strand can
bind to another upper strand because their primer portions are not complementary. For
the same reason, two lower strands cannot bind. Thus, we compose a selection matrix by
counting mishybridized bases of upper strand i and lower strand j when these strands
have their (fully complementary) primer sequences aligned. An example of four
mishybridized bases can have the form:
|
S1 |
| S2
|
MMM...MMMACGTACGT...ACGTACGTMMM...MMM -> 3’
3’ <- NNN...NNNCATGTGCA...TGCATGCANNN...NNN
|
S1’ |^^^^
| S2’ |
||||
mishybridizations
An alternative way of counting the number of mishybridized bases complements the
bases in the lower strand, except those in S1’ and S2’. The equivalent computation is:
|
S1 |
| S2
|
MMM...MMMACGTACGT...ACGTACGTMMM...MMM -> 3’
3’ <- NNN...NNNGTACACGT...ACGTACGTNNN...NNN
|
S1’ |^^^^
| S2’ |
||||
disagreements
Thus, the count of the number of disagreeing bases in an upper strand and a lower strand
(disregarding the primer parts) equals the number of mismatched bases in the paring of
the upper strand to the base-by-base complement of the lower strand. The count of
disagreeing symbols in two sequencesof equal length is known as the Hamming distance
between the two sequences.
In summary, to form a simplified selection matrix, first let the distinct DNA
sequences be assigned a specific (but arbitrary) order. Second, ignore the primer parts of
the DNA sequences. Third, assign 1 to the i,j element of the selection matrix $S$ if the
Hamming distance between the ith sequence and the jth sequence exceeds a specified
threshold, and assign zero otherwise.
If only two of the four bases are used---for example A and T might be identified
with the binary digits 0 and 1--and all possible binary sequences of a fixed length are
arranged in numerical order, then the resulting selection matrix is a focus in research in
binary error-correcting codes. The temperature selection protocol, in seeking a maximal
set of non-crosshybridizing DNA sequences of A and T, is seeking a maximum size
binary error-correcting code. This is also true when using three of the four bases or all
four bases. However, as mentioned before, the results may be suggestive, but it is
problematic to merely count mismatches to estimate the likelihood of hybridization
except for A and T only.
The link between non-crosshybridizing libraries of DNA strands and errorcorrecting codes is valuable. For one thing, various upper and lower bounds are known
for the maximum size of error-correcting codes. Also, evolutionary computational
techniques for finding maximum error-correcting codes can be compared to the
laboratory evolution of DNA computing libraries.
Examples: Binary Sequences of Length Two
Important concerns about temperature selection PCR can be illustrated with small
examples of our model. Consider the four binary sequences of length 2: [00, 01, 10, 11],
or [AA, AT, TA,TT]. The matrix of Hamming distances is
0
1
1
2
1
0
2
1
1
2
0
1
2
1
1
0
Distances exceeding 1 give the selection matrix
0
0
0
1
0
0
1
0
0
1
0
0
1
0
0
0
One maximum library has upper strands {AA, TT}. This library cannot be enlarged by
including either AT or TA because they would have only one mismatch with the other
library sequences. A second library is {AT, TA}.