Systems Biotechnology Biotechnology and Bioengineering
advertisement

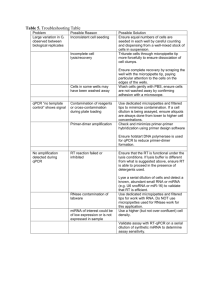
Supplementary Figure 1: cgr-miR-185-5p is a stable expressed miRNA under various conditions and a suitable “housekeeping RNA” for normalization of qPCR data a) Transcript levels of mature cgr-miR-185-5p during batch cultivation of four different CHO-K1 cell lines [1]. Expression levels are normalized to early exponential growth phase at day 0, and show only minor changes (log2 < 0.3) during late exponential (day4) or stationary/early decline (day8) phase. b) Expression levels of miR-185-5p and miR-185-3p were measured by nextgeneration sequencing in 6 distinct CHO cell lines [2]. c and d) Comparison of log2 fold changes for 9 genes (4 samples each) measured by microarray (y-axis) and qPCR (x-axis). qPCR data were normalized either using miR-185-5p or Glycerin-aldehyde-3-phosphateDehydrogenase (gapdh) as internal control. Pearson correlations of 0.67 (gapdh) and 0.93 (miR-185-5p) indicate that miR-185-5p is equally well suited as internal control as gapdh. Supplementary Figure 2: Outline of screening method 1. CHO-EpoFc producer cell line was transfected with miRNA expression vector. 2 the transfected cells were transferred to conical shaker flask to start batch culture. 3 Effect of miRNA overexpression was observed on growth, viability and productivity. The outlined method can be a general platform for screening for microRNAs, miR-sponges, siRNAs or mRNA overexpression along with detailed functional characterization. Supplementary Figure 2. cgr-miR-185-5p is a stable expressed miRNA under various conditions and a suitable “housekeeping RNA” for normalization of qPCR data a) Transcript levels of mature cgr-miR-185-5p during batch cultivation of four different CHO-K1 cell lines [1]. Expression levels are normalized to early exponential growth phase at day 0, and show only minor changes (log2 < 0.3) during late exponential (day4) or stationary/early decline (day8) phase. b) Expression levels of miR-185-5p and miR-185-3p were measured by nextgeneration sequencing in 6 distinct CHO cell lines [2]. c and d) Comparison of log2 fold changes for 9 genes (4 samples each) measured by microarray (y-axis) and qPCR (x-axis). qPCR data were normalized either using miR-185-5p or Glycerin-aldehyde-3-phosphateDehydrogenase (gapdh) as internal control. Pearson correlations of 0.67 (gapdh) and 0.93 (miR-185-5p) indicate that miR-185-5p is equally well suited as internal control as gapdh. [1] J.A.H. Bort, M. Hackl, H. Höflmayer, V. Jadhav, E. Harreither, N. Kumar, et al., Dynamic mRNA and miRNA profiling of CHO-K1 suspension cell cultures, Biotechnol J. (2011). [2] M. Hackl, T. Jakobi, J. Blom, D. Doppmeier, K. Brinkrolf, R. Szczepanowski, et al., Nextgeneration sequencing of the Chinese hamster ovary microRNA transcriptome: Identification, annotation and profiling of microRNAs as targets for cellular engineering, J. Biotechnol. 153 (2011) 62-75. Supplementary Figure 2. Outline of screening method 1. CHO-EpoFc producer cells ere transfected with miRNA expression vector 2. transfected cells were transferred to conical shaker flask to start batch culture 3. Effect of miRNA overexpression was observed on growth, viability and productivity. The outlined method can be a general platform for screening of effects of microRNAs, miRsponges, siRNAs or mRNA overexpression along with detailed functional characterization.