Figure S1. Immunoblot analysis of Rbr2 content in D. vulgaris strains
advertisement

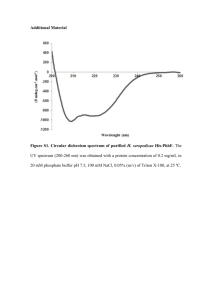
1 2 3 4 5 6 7 97.0 kDa 67.0 kDa 43.0 kDa 1 2 3 4 5 6 7 37.1 kDa 30.0 kDa 25.9 kDa 19.4 kDa 20.1 kDa 14.8 kDa 14.4 kDa A B Figure S1. Immunoblot analysis of Rbr2 content in D. vulgaris strains. (A) Coomassie Blue stained gel following SDS-PAGE. Lane 1: 70 μl of wild-type strain protein extract [cells from 7 ml of culture (OD 600 = 0.356) were pelleted, resuspended and boiled for 2 min in 250 μl of IBuffer and 250 μl of water]; lane 2: 30 μl of wild-type strain protein extract; lane 3: 70 μl of D. vulgaris rbr2 protein extract [cells from 7 ml of culture (OD600 = 0.413) were pelleted, resuspended and boiled for 2 min in 250 μl of IBuffer and 250 μl of water]; lane 4: 30 μl of D. vulgaris rbr2 extract; lane 5: 5 μg of purified Rbr2 protein; lane 6: 3.3 μg of purified Rbr2 protein; lane 7: LMW protein markers. (B) Immunoblot of the gel as in (A), except that BenchMark prestained markers were used. Experimental details: The QIAexpress System (QIAGEN) was used to overexpress Rbr2. Primers rbr2-f (ggggatccATGTCCA AGACCCATGAGAAC) and rbr2-r (ggggtaccCTAGTCGACCTTGTAGAAGGC) were used to amplify the rbr2 gene of D. vulgaris from 100 ng of genomic DNA. Using the QIAquick PCR purification kit (QIAGEN), 100 μl of PCR product was cleaned and then digested with KpnI and BamHI. The digested fragment was gel purified and then ligated to similarly digested and gel-purified pQE30 (QIAGEN). Plasmid, pQE30-rbr2 was then transformed into E. coli M15 [pREP4] in order to overexpress Rbr2. 1000 ml of TY containing 100 μg/ml of ampicillin and 25 μg/ml of kanamycin was inoculated with 20 ml of an overnight culture of E. coli M15 [pREP4/pQE30-rbr2] and grown to an OD600 of around 0.6 by shaking ~200 rpm at 37C. The expression of Rbr2 was then induced by the addition of 1 mM of IPTG followed by growth for an additional 5 h. Cells were harvested by centrifugation at 7000 rpm in a Sorvall RC-5B centrifuge for 20 min, resuspended in Buffer B (100 mM NaH2PO4, 10 mM Tris-HCl, 8 M urea; pH 8.0) at 5 ml per gram wet weight and then frozen at -70C. The overexpressed Rbr2 was purified with Ni-NTA agarose according to the manufacturer’s instructions. Cells were lysed with a French Press (1000 psi; twice) and the lysate was centrifuged in a Sorvall RC-5B centrifuge for 20 min at 10000 rpm to pellet the cellular debris. Supernatant (4 ml) was then mixed with 1 ml of 50% Ni-NTA slurry, equilibrated with Buffer B, followed by incubation with gentle shaking for 20 min. The lysate-resin mixture was loaded into a column. The flow-through was collected and the column was washed twice with 4 ml of Buffer C (100 mM NaH2PO4, 10 mM Tris-HCl, 8 M urea; pH 6.3) and the wash fractions were collected. The overexpressed Rbr2 was then eluted with 4 x 0.5 ml of Buffer D (100 mM NaH2PO4, 10 mM Tris-HCl, 8 M urea; pH 5.9 with HCl). The presence of essentially pure Rbr2 in all fractions was indicated by SDS-PAGE. Fractions were dialyzed against TrisEDTA (pH 7.4) at 4C overnight to remove SDS and then against PBS (0.14 M NaCl, 2.7 mM KCl, 10 mM Na 2HPO4, 1.5 mM KH2PO4; pH 7.2), giving a solution of 325 μg/ml of Rbr2 in PBS. Polyclonal antibodies, recognizing D. vulgaris Rbr2, were prepared by immunizing a rabbit with 325 μg of purified Rbr2 in PBS and giving a booster shot 4 weeks later with 220 μg of purified Rbr2. Blood was collected 10 days after the booster shot and prepared serum was stored frozen at -70C. SDS-PAGE was carried essentially according to Laemmli (1970. Nature. 227: 680-685). Proteins were then electroblotted onto a nitrocellulose membrane as described by Towbin et al., (1979. Proc. Natl. Acad. Sci. U S A. 76: 4350-4354). Gelatin-blocked nitrocellulose filters were incubated with a 1000-fold diluted primary antibody. The latter was deteced by incubating with 2000-fold diluted alkaline phosphatase-conjugated secondary antibody.