PPM1D OVEREXPRESSION AND GENE AMPLIFICATION IN
advertisement

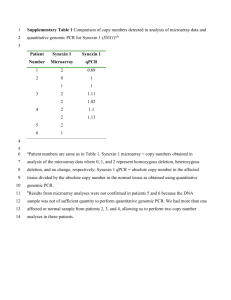
1 1 Supplementary Materials Supplementary Table 1: Summary of antibodies, clones, dilutions, antigen retrieval methods and scoring systems. Supplementary Figure 1: Representative genome plots of grade III invasive ductal carcinomas of no special type harbouring PPM1D gene amplification (17q23.2) obtained with a 32K tiling path bacterial artificial chromosome (BAC) microarray comparative genomic hybridisation platform and previously reported by Natrajan et al. . Please note that all cases displayed clustered regions of amplification mapping to a given chromosomal arm and multiple regions of amplification throughout the genome (A-H), consistent with the definition of a ‘firestorm’ profile as defined by Hicks et al. . In A to H, Log2 ratios are plotted on the Y axis against each BAC clone according to genomic location on the X axis. BACs categorised as displaying genomic low level gains as defined by circular binary segmentation-smoothed (cbs) ratios > 0.12 are highlighted in dark green, those categorised as amplification as defined by cbs-smoothed ratios > 0.45 are highlighted in bright green, and those categorised as genomic losses as defined by cbs-smoothed ratios < -0.12 are highlighted in red. Details of the microarray hybridisations, image capturing and data analysis are described elsewhere .