pbi12073-sup-0003-TextS1
advertisement

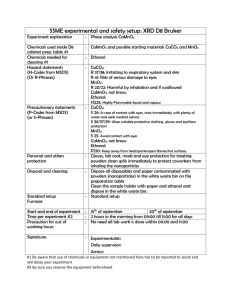
SUPPLEMENTRARY INFORMATION Marker-free genetic engineering of the chloroplast in the green microalgae Chlamydomonas reinhardtii Running title: Marker-free microalgal chloroplast transformation Hsu-Ching Chen and Anastasios Melis* Department of Plant and Microbial Biology 111 Koshland Hall, MC-3102 University of California Berkeley, CA 94720, USA * Author for correspondence Email: melis@berkeley.edu; Fax: 510-642-8166 Strains, culture media, and growth conditions Strain CC2653 is deficient in RuBisCO and unable to grow photo-autotrophically. It requires acetate for growth, and is light sensitive. This strain was cultivated in a TRIS–Acetate– Phosphate (TAP) medium, pH 7.0 (Gorman and Levine 1965) under dim light or in the dark. Other Chlamydomonas strains were grown either photo-mixotrophically in TAP medium, or photo-autotrophically in HS minimal medium (Sueoka 1960). Cells in liquid culture were grown in Erlenmeyer flasks at 24C with shaking in the dark or under continuous illumination at approximately 50 µmol photons m-2 s-1, or under 12-h light-12-h dark incubation. Culture density was measured either by cell counting using a Neubauer ultraplane hemacytometer and a BH-2 light microscope (Olympus, Tokyo), or from the light scattering of an aliquot of the 1 culture at 730 nm using a UV-VIS spectrophotometer (Shimadzu UV1800, Tokyo). Extraction of genomic DNA Total DNA was extracted from Chlamydomonas reinhardtii cells by using a hexadecyltrimethylammonium bromide (CTAB) based method. Cells were collected from 200 ml of late log phase cultures and resuspended in 5 ml of 2% CTAB extraction buffer containing 1.4 M NaCl, 100 mM Tris.HCl pH 8.0, 20 mM EDTA. Proteinase K was added to a final concentration of 0.5 mg/ml, then the mixture was incubated at 65°C for 90 min. At the end of incubation, 50 µl RNaseA (10 mg/ml) was added and incubation was extended for 30 min at room temperature. At the end of incubation, suspension was centrifuged at 5,200 g for 5 min to remove cell debris. The supernatant was then extracted with chloroform:isoamyl-alcohol (24:1), followed by isopropanol precipitation. Southern blot analysis was carried out by using Amersham AlkPhos Direct™ Labeling and Detection Systems (GE Healthcare, USA) according to the manufacturer’s instructions. Expression vector for Chlamydomonas reinhardtii chloroplast transformation The alignment of the nucleotide sequence from the Saccharomyces cerevisiae ADH1 gene (ScADH1) and the corresponding codon-optimized gene for Chlamydomonas reinhardtii chloroplast transformation (CrCpADH1) is shown in supplementary Figure S1. Generation of CrCpADH1 specific polyclonal antibodies The coding sequence of the CrCpADH1 gene, with or without the 90 bp of rbcL 5’ coding region, was cloned into pET vector for over-expression of the protein. The expressed CrCpADH1 protein was purified from E. coli using Ni-NTA agarose column according to manufacturer’s instructions (Qiagen, Valencia, CA), and used as antigen for the generation of specific polyclonal antibodies (Covance, Denver, PA ). Two constructs were made for their expression in E. coli: one contains an ORF (open reading frame) with the extra 90 bp from rbcL coding sequence which corresponds exactly to the one we introduced into Chlamydomonas chloroplast, the other contains only the coding region of the CrCpADH1 without the rbcL 90 bp sequence. 2 Supplementary Figure S2(a) shows the over-expression profiles of these two types of proteins. High levels of CrCpADH1 expression were obtained in E. coli with both constructs upon overnight induction. Also, among 12 randomly selected clones from the rbcL90-CrCpADH1 overexpression strains, we have detected only one type of protein of which the size corresponds to the theoretical size of rbcL90-CrCpADH1, indicating that there is no translation initiation from the second ATG (the initiation codon of CrCpADH1 gene). Fig. S2(b) shows the purified CrCpADH1 protein after Ni-NTA column affinity chromatography. The purified CrCpADH1 protein was used as antigen for the generation of specific polyclonal antibodies. Protein analysis by SDS-PAGE and Western blotting For total protein extraction, cells were harvested by centrifugation at 3,000 g for 5 min at 4°C, and cell pellets were resuspended in ice-cold sonication buffer containing 50 mM Tricine (pH 7.8), 10 mM NaCl, 5 mM MgCl2, 0.2% polyvinylpyrrolidone-40, 0.2% sodium ascorbate, 1 mM aminocaproic acid, 1 mM aminobenzamidine and 100 µM phenylmethylsulfonylfluoride (PMSF). Cells were broken by sonication in a Branson 250 Cell Disruptor operated at 4°C for 30 s (pulse mode, 50% duty cycle, output power 5). An equal volume of 2x protein solubilization buffer containing 0.5 M Tris-HCl (pH 6.8), 7% SDS, 20% glycerol, 2 M urea, and 10% mercaptoethanol was added immediately prior to SDS-PAGE analysis. In the case of smaller cell volumes, cell pellets were re-suspended directly in 1x protein solubilization buffer on the basis of equal chlorophyll concentration and incubated at room temperature for 2-3 h. Chlorophyll content was determined from the absorbance of a pigment extract in 80% acetone spectrophotometrically (Arnon 1949). Solubilized protein extracts were resolved by SDS-PAGE using the discontinuous buffer system of Laemmli (1970). After completion of PAGE, proteins were either stained with Coomassie Brilliant Blue or electro-transferred onto PVDF membrane. Immunoblot analysis was carried out with specific polyclonal or monoclonal antibodies, followed by chemiluminescence detection of the cross reactions using the SuperSignal West Pico Chemiluminesencet substrate (Pierce-Thermo Scientific). 3 Ethanol production measurements Cells were grown to the mid exponential growth phase (about 4-5x106 cells/ml) in Erlenmeyer flasks at 23C upon orbital shaking under 12 h-light – 12-h dark cycles with a light intensity of 50 µmol photons m-2 s-1. Cultures were then centrifuged and cell pellet was resuspended in TAP-S medium at a chlorophyll concentration of ~ 50 µg/ml. Cells were then incubated in the dark up to a total period of 120 h, with samples taken at 24 h intervals for analysis. The ethanol content of the liquid medium in the culture was quantified using a calibrated GC-FID analysis (Shimadzu, G8A). Briefly, an aliquot of 100 µl of cell suspension was placed in 2 ml glass vials, sealed with rubber stoppers, incubated at 55°C for 20 min, then 1 ml of gas sample was taken from the headspace of the glass vial and injected into GC-FID. Ethanol concentration calibration curves were also obtained through GC-FID measurements by using this method (Fig. S3(a)). A linear relationship between ethanol concentration and the GC-FID amplitude of the ethanol peak with a retention time of 1.5 min was established with ethanol concentrations ranging from 0.01% to 0.1% v/v (Fig. S3(b)). Supplementary References Arnon, D. (1949) Copper enzymes in isolated chloroplasts. Polyphenol oxidase in Beta vulgaris. Plant Physiol 24: 1–15 Gorman, D. S., and Levine, R. P. (1965) Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl Acad. Sci. USA 54, 1665-1669. Laemmli, U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 Sueoka, N. (1960) Mitotic Replication of Deoxyribonucleic Acid in Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 46, 83-91. 4 Legends to Supplementary Figures Figure S1. Sequence alignment of ADH1 gene from Saccharomyces cerevisiae (ScADH1) and the Chlamydomonas reinhardtii chloroplast codon optimized ADH1 gene (CrCpADH1). Translation initiation codon ATG and termination codon TAA are indicated in red- font underlined. The 6xHis tag at the 3’ end of the CrCpADH1 gene is also underlined. Figure S2. Expression of rbcL90-CrCpADH1 and CrCpADH1 proteins in E. coli. (a) SDS-PAGE analysis of total protein extracts from E. coli. 1-3 are protein extracts from the same rbcL90CrCpADH1 over expressing strain but under three different treatments: Lane 1: 1-h induction; Lane 2: no-induction and Lane 3: overnight induction. Samples from lanes 4 & 5 are overnight induction of two CrCpADH1 over expressing strains. (b) Purified CrCpADH1 protein after Ni-NTA column affinity chromatography. Lanes 1 & 2 represent products from two independent purifications. Figure S3. Ethanol production measurements. (a) GC-FID chromatograms of ethanol standard solutions measured with a GC-FID Shimadzu 8A apparatus. (b) Ethanol concentration standard calibration curve in the concentration range of 0 to 0.1% (v/v). 5