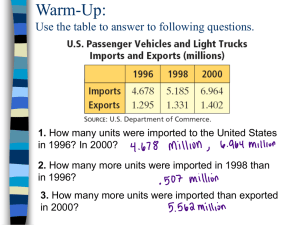
Derivation of the BLOSUM substitution matrices:
advertisement

Bioc 565, Dr. Cordes Supplement handout to sequence evolution lecture, Sep 10, 2007 1. General facts about amino-acid substitution matrices: The average likelihood of a particular mutation occurring and being accepted is influenced both by codon similarity and by chemical similarity of the amino acids (as a result of structural and functional constraints). These likelihoods are reflected in substitution matrices. The most common methods for constructing substitution matrices are empirical, i.e. the matrices are parametrized using observations of what substitutions actually occur among collections of related protein sequences. Substitution matrices are today commonly used in sequence alignment methods and database sequence similarity searches. A pioneer in this field was Margaret Dayhoff, who created the PAM (Per cent Accepted point Mutations) substitution matrices. She used sets of closely related sequences to model how evolution occurs over short distances, and then extrapolated these findings to longer evolutionary distances. The other most commonly used type of substitution matrix is called the BLOSUM matrix, due to Henikoff. BLOSUM matrices use less closely related sequences for parametrization, and only use “blocks” of the best aligned portions. Portions that don’t align well and vary in length, such as surface loops, are not used. Thus the PAM and BLOSUM matrices show some differences. Matrices parametrized using soluble, globular proteins will also differ from those derived from transmembrane proteins. A few words on nomenclature: you will see terms like “BLOSUM 62” and “PAM 250”. “BLOSUM 62” means that the matrix was parametrized using sequences with 62% average sequence identity. “PAM 250” refers to the PAM unit: 1 PAM is the amount of evolution required to change 1% of a protein’s residues. A PAM 250 matrix is then the result of extrapolating data on closely related sequences to an evolutionary distance of 250 PAMs. Another thing you will note is that the numbers in the PAM matrix don’t look like “percent accepted” values--many are negative numbers. This is because most matrices are presented in “log odds” form. The odds are figured as the probability of a particular change at a given evolutionary distance, normalized by the overall frequency of occurrence of a given amino acid in the database. These odds are then converted to logarithms. page 1 of 4 2. Derivation of substitution matrices from sequence conservation data: Construction of the BLOSUM matrices based on Henikoff & Henikoff, PNAS USA, 89, 10915 (1992) The general procedure for deriving generalized substitution matrices from sequence conservation data is to 1) assemble a large number of multiple alignments from different protein families 2) tabulate the frequencies of all the possible residue pairings 3) compare these frequencies to those expected in a random database of the same composition, giving an odds ratio 4) convert the odds ratio into a log format. To make the BLOSUM matrices (which are probably the best and are certainly the most commonly used), Henikoff and Henikoff took multiple alignments of several hundred protein families and parsed them into a database of “blocks”, where a block is defined as a portion of a multiple alignment which contains no gaps. This analysis yielded a database of >2000 blocks. The block approach contrasts with the way the PAM matrices (Dayhoff) were constructed, because in that case regions containing gaps were included. There are 20 different naturally occurring amino acids, and thus there are 20 + 19 + ... 1 = 210 different possible pairings of amino acids. Henikoff and Henikoff converted their database of blocks into a frequency table describing the number of occurrences fij of each of the 210 possible pairings of amino acids i and j, where 1 ≤ j ≤ i ≤ 20. To illustrate construction of a frequency table, let’s look at a simple (and fake) example. A given block can be described as having a width of w alignment positions and a depth of s sequences. The very small block shown below has w=4 (positions 1-4) and s=5 (sequences A-E). Let’s suppose that our entire database is composed of this one block. sequence A B C D E position in alignment 1 2 3 4 S L M K A L A E M V A E R I A W T L M C A block of w=4 and s=5 has a total of ws(s-1)/2 = 40 amino acid pairs. Let’s now consider, as an illustrative exercise, the frequencies of all pairs involving alanine. Alanine occurs at two positions in the block: positions 1 and 3. At the first position, there are 4 alanine-containing pairs: 1 each of the pairs AM, AR, AS and AT. At the third position, there are 9 alanine-containing pairs, including 3 different AA pairs (sequences B and C, B and D and C and D) and 6 AM pairs page 2 of 4 (enumerate them yourself to convince yourself). So summing the occurrences of each pair at the two positions, the frequency of occurrence of AM pairs fAM = 1+ 6 = 7, the frequency of occurrence of AA pairs fAA = 0 + 3 = 3. The frequency of occurrence of AR, AS and AT pairs is just 1, while pairings of alanine with all other amino acids have frequencies of 0. If we did this same analysis with all 210 possible pairings we’d have a pair frequency table (or matrix) for our database. The pair frequencies are then converted into pair probabilities. The probability of observing any particular pairing can be described as The denominator here simply corresponds to the sum total of all pair frequencies (which is just the total number of pairs) in the database. qAM for example, is 7/40 = 0.175, while qAA is 3/40 = 0.075. The pair probabilities only have statistical meaning when compared to the pair probabilities that one would expect from a random database of the same aminoacid composition. The expected probability of occurrence eij for each pair, based on the frequency of occurrence of pairs involving each of those amino acids in the database, is equal to pipj, where pi and pj are the individual probabilities that a given residue pair will contain amino acid i or j, respectively. The probability pi that a pair will contain amino acid i is generally figured as: I find this formulation mildly confusing, and it should be noted that this number will also come out to equal the fractional population of amino acid i in the database. This makes sense. Intuitively, the probability of a given pair containing alanine should be equal to the fractional population of alanine in the database. For example, as figured from the equation above, pA = [3 + (10/2)]/40 = 0.2. The fractional population of alanine in the database is also 4/20 = 0.2. Similarly, pM = [1 + (10/2)]/40 = 0.15. The fractional population of methionine in the database is 3/20 = 0.15. In any case, for the example of alanine-methionine pairs, eAM = (0.2)(0.15) = 0.03. Similarly, for alanine pairing with itself, eAA = (0.2)(0.2) = 0.04. The odds ratio is then computed as the observed probability qj divided by the expected probability eij . This ratio is then converted into the log-odds ratio sij, where sij = log2(qij/eij). page 3 of 4 For the AM pairing, qAM/eAM = 0.175/0.03 = 5.82, and sAM = 2.54. For the AA pairing, qAM/eAM = 0.075/0.04 = 1.88, and sAM = 0.9. Note that if qij is larger than eij, the odds ratio will be greater than 1 and the log-odds score will be positive, while if the opposite is true the odds ratio will be less than 1 and the log-odds score will be negative. Thus, pairings which occur more frequently than expected by chance will have positive log odds scores, while those which occur less frequently will have negative log odds scores. Note that this particular case is unrealistic: in any real situation, alanine would have a higher log odds score with itself than with any other amino acid. To convert them to final BLOSUM matrix element form, the log odds scores sij are then all multiplied by some uniform scaling factor (2 in the case of the BLOSUM matrices) and rounded off to integers. page 4 of 4