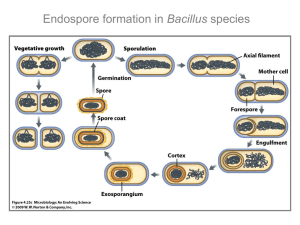
Daptomycin versus Friulimicin:
advertisement

1 DETAILED MATERIAL AND METHODS FOR WECKE ET AL. 2008 2 Daptomycin versus Friulimicin B: In-depth Profiling 3 of the Bacillus subtilis Cell Envelope Stress Responses 4 5 Strains, media and growth conditions. B. subtilis strains were routinely grown in LB medium 6 at 37C with aeration, except where stated otherwise. Erythromycin (1 g/ml) plus lincomycin (25 7 g/ml) for MLS resistance were used for the selection of strain BFS2469, tetracycline (10 µg/ml) was 8 used for selection of strain TMB389. FRI was obtained from MerLion Pharmaceuticals GmbH, and 9 other drugs from their respective manufacturers. 10 RNA preparation. B. subtilis 168 wild type strain was grown aerobically at 37°C in LB medium 11 to mid-log phase. The culture was split and induced with FRI or DAP (1 µg/ml each) with one sample 12 remaining as the uninduced control. After 10 min of induction 30 ml of each sample were mixed with 13 15 ml cold killing buffer (20 mM Tris-HCl pH 7.0, 5 mM MgCl2, 20 mM NaN3), harvested by 14 centrifugation and frozen in liquid nitrogen. For cell disruption, the pellet was resuspended in 200 µl 15 killing buffer, immediately dropped into the Teflon vessel (filled and pre-cooled with liquid nitrogen), 16 and then disrupted with a Mikro-Dismembrator U (Sartorius). The resulting cell powder was 17 resuspended in 3 ml of lysis solution (4 M guanidine-thiocyanate, 0.025 M Na-acetat pH 5.2, 0.5% N- 18 lauroylsarcosinate) and the RNA was extracted twice by phenol/chloroform/isoamylalcohol 25/24/1 19 followed by chloroform/isoamylalcohol 24/1 extraction and ethanol precipitation. RNA samples were 20 DNase-treated with the RNase-free DNase kit (Qiagen) according to the manufacturer’s instructions 21 and purified using RNeasy mini columns (Qiagen). The quality control of the RNA preparations was 22 performed with the RNA 6000 Nano LabChip Kit (Agilent Technologies) on the Agilent 2100 23 Bioanalyzer according to the manufacturer’s instructions. 24 DNA microarray analysis. The RNA samples obtained from three independent cultivations 25 were used for independent cDNA synthesis and DNA array hybridization. Generation of the Cy3/Cy5- 26 labeled cDNAs and hybridization to B. subtilis whole-genome DNA microarrays (Eurogentec) were 27 performed as described (6). The slides were scanned with a ScanArray Express scanner 28 (PerkinElmer). Quantitation of the signal and background intensities was carried out using the 29 ScanArray Express image analysis software. 30 Transcriptome data analysis. Data was analyzed using the GeneSpring software (Agilent 31 Technologies). Raw signal intensities were first transformed by intensity dependent LOWESS 32 normalization. The normalized array data were subjected to a statistical analysis using Cyber-T, a 33 program based on a t-test combined with a Bayesian statistical framework (2). The software is 34 accessible through a web interface at http://cybert.microarray.ics.uci.edu. The mRNA abundance was 35 considered to be significantly different between the untreated control samples and the samples 36 obtained after treatment with the respective antibiotic if (i) the Cyber-T Bayesian P value was < 0.001 37 and (ii) the average fold change was at least 3 in three independent experiments. The potential and 38 known functions of the encoded proteins 1 were initially inferred from the SubtiList 1 (http://genolist.pasteur.fr/SubtiList/) or BSORF databases (http://bacillus.genome. ad.jp/). An in-depth 2 analysis of the identified marker genes (as listed in Table 2) was performed using the SMART (7, 9) 3 and 4 http://www.microbesonline.org/, respectively. 5 MicrobesOnline (1) databases, at http://smart.embl-heidelberg.de/ and Quantitative real time RT-PCR. Measurement of transcript abundance was performed by 6 quantitative real-time RT-PCR using iScript one-step RT-PCR kit with SYBR Green (Bio-Rad) 7 according to the manufacturer’s procedure with minor modifications: In brief, 100 ng of DNA-free total 8 RNA was used in a total reaction volume of 20 µl with 0.3 µM of each primer (see Table 1). The 9 amplification reaction was carried out in an MyiQ Cycler (BioRad) using the following program: reverse 10 transcription at 50°C for 10 min, followed by a 95°C denaturing/activation step for 5 min, followed by 11 45 cycles (95°C for 10 sec), (60°C for 30 sec). After a subsequent denaturation (95°C for 1 min) and 12 annealing (55°C for 1 min) the setpoint temperature was increased in 80 cycles (10 sec each) by 13 0.5°C/cycle, starting from 55°C, to determine the melting temperatures of the PCR products. 14 Expression of rpsJ and rpsE was monitored as constitutive reference. These genes were chosen due 15 to their stable expression behaviour under various growth and stress conditions in B. subtilis (data not 16 shown). Expression of liaI and genes encoding ECF factors was calculated as fold changes using 17 the formula: Fold change = 2-Ct; with -Ct = (Ct(gene x)-Ct(constitutive gene))condition I – (Ct(gene x)-Ct(constitutive 18 gene))condition II (11). Concentration-dependent killing curve/β-galactosidase assays. These experiments were 19 20 performed as described (8). In brief, strain BFS2470 was grown in LB medium with MLS selection to 21 OD600 ~ 0.5 and DAP/FRI were added to a final concentration ranging from 0.01 to 50 g/ml. An 22 uninduced culture was used as a negative control. The cultures were incubated with aeration at 37°C. 23 A sample was taken after 30 min for -galactosidase assay and the turbidity of the remaining culture 24 was measured for at least 5 hours to monitor the concentration-dependent effects of the antibiotics on 25 growth. 26 L-[35S]methionine labelling of proteins and 2D-PAGE analysis. B. subtilis 168 wild type 27 strain was grown aerobically at 37°C in Belitsky minimal medium (10) to mid-log phase. 10 and 30 28 minutes after addition of DAP or FRI (1,5 and 1,0 µg/ml, respectively) the cells as well as untreated 29 control cells were labelled with 15 µCi/ml [35S]-methionine. After 5 minutes incorporation of radioactive 30 methionine, the reaction was stopped by adding an excess of nonradioactive methionine (1mM) and 31 chloramphenicol (100µg/ml) to stop translation. Samples were taken and the cytoplasmic protein 32 fraction isolated as described earlier (3). 2D-PAGE using Immobiline dry strips (IPG, Amersham 33 Biosciences) (pH 4-7) loaded with 80 µg protein extract as well as visualization of radiolabelled 34 proteins and dual channel imaging using Delta2D software (Decodon) were carried out as described 35 (4, 5). 36 2 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 REFERENCES 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. Alm, E. J., K. H. Huang, M. N. Price, R. P. Koche, K. Keller, I. L. Dubchak, and A. P. Arkin. 2005. The MicrobesOnline web site for comparative genomics. Genome Res 15:1015-22. Baldi, P., and A. D. Long. 2001. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17:509-19. Bandow, J. E., H. Brotz, L. I. Leichert, H. Labischinski, and M. Hecker. 2003. Proteomic approach to understanding antibiotic action. Antimicrob Agents Chemother 47:948-55. Bernhardt, J., K. Buttner, C. Scharf, and M. Hecker. 1999. Dual channel imaging of two-dimensional electropherograms in Bacillus subtilis. Electrophoresis 20:2225-40. Bernhardt, J., J. Weibezahn, C. Scharf, and M. Hecker. 2003. Bacillus subtilis during feast and famine: visualization of the overall regulation of protein synthesis during glucose starvation by proteome analysis. Genome Res 13:224-37. Jürgen, B., S. Tobisch, M. Wümpelmann, D. Gördes, A. Koch, K. Thurow, D. Albrecht, M. Hecker, and T. Schweder. 2005. Global expression profiling of Bacillus subtilis cells during industrial-close fed-batch fermentations with different nitrogen sources. Biotechnol Bioeng 92:277-98. Letunic, I., R. R. Copley, B. Pils, S. Pinkert, J. Schultz, and P. Bork. 2006. SMART 5: domains in the context of genomes and networks. Nucl. Acids Res. 34:D257-260. Mascher, T., S. L. Zimmer, T. A. Smith, and J. D. Helmann. 2004. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob Agents Chemother 48:2888-96. Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A 95:5857-64. Stülke, J., R. Hanschke, and M. Hecker. 1993. Temporal activation of beta-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J Gen Microbiol 139:2041-5. Talaat, A. M., S. T. Howard, W. Hale IV, R. Lyons, H. Garner, and S. A. Johnston. 2002. Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucl. Acids Res. 30:e104-e109. 3