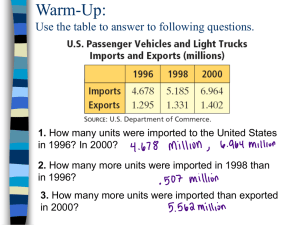
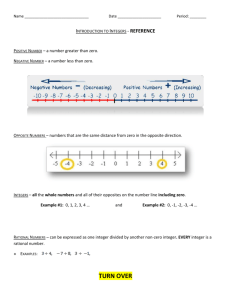
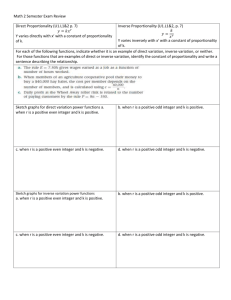
Copyright 1990--2002 Gerald Z. Hertz May be copied for
advertisement

Copyright 1990--2002 Gerald Z. Hertz May be copied for noncommercial purposes. Author: Gerald Z. Hertz gzhertz AT alum.mit.edu CONSENSUS (version 6d) REQUIRED PARAMETER -L <Width of pattern (required)> BASIC OPTIONS -h <Print directions> -f <Name of sequence file> -a <Name of ascii alphabet file (default: alphabet)> -A <Ascii alphabet information on the command line> -d <Use designated prior frequencies (default: use observed frequencies)> -c0 <Ignore the complementary strand (the default)> -c1 <Include both strands as separate sequences> -c2 <Include both strands as a single sequence (i.e., orientation unknown)> -c3 <Assume that the pattern is symmetrical> -l <Seed with first sequence and proceed linearly through list> -n <Maximum number of cycles (0 or more sites per sequence)> -N <Maximum number of cycles (1 or more sites per sequence)> ADVANCED OPTIONS -q <Number of matrices to save (default: 1000)> -m <Minimum distance between words (default: integer indicated by -L option)> -t <Terminate indicated number of cycles after most significant alignment> -pt <Number of top matrices to print (default: 4 when NOT using -l option)> -pf <Number of final matrices to print (default: 4 when NOT using -n or N)> -u0 <Unrecognized characters are errors> -u1 <Unrecognized characters are discontinuities, but print warning (default)> -u2 <Unrecognized characters are discontinuities, and print NO warning> OBSCURE OPTIONS -i <Name of integer alphabet file> -CS <Ascii alphabet is case sensitive (default: case insensitive)> -w <Only count letters included within the sequence fragments being aligned> -pr1 <Save top progeny matrices regardless of parentage> -pr2 <Save the top progeny for each parental matrix (the default)> This program determines consensus patterns in unaligned sequences. The algorithm is based on a matrix representation of a consensus pattern. Each row corresponds to one of the letters of the relevant alphabet---e.g., 4 rows in the case of DNA. Each column corresponds to one of the positions within the pattern. The elements of the matrix are determined by the number of times that the indicated letter occurs at the indicated position. Matrices are constructed by sequentially adding additional L-mers (subsequences of length L, where L is the width of the pattern being sought) to previously saved matrices. During each cycle, only the most significant matrices are saved. The maximum number of matrices to save is determined by the "-q" option (see section 1 below). In practice, less matrices are ultimately saved because many of the matrices initially saved are identical to each other. The program can use 3 different criteria for deciding to stop adding additional words to the saved matrices: 1) Each sequence has contributed exactly one word to the saved matrices (the default). 2) The saved matrices contain a maximum allowable number of words (set with the "-n" and "-N" options). 3) The program has completed a designated number of cycles since finding the current most significant alignment (set with the "-t" option). This latter criteria is used in addition to criteria 1 and 2 to terminate the program sooner. The significance of a matrix is initially measured by its information content. A higher information content indicates a rarer pattern and a more desirable matrix. The program also estimates for each matrix a p-value, which is the probability of observing the particular information content or higher in an arbitrary alignment of random L-mers. The ultimate statistical significance of a matrix is determined by multiplying the p-value by the approximate number of possible alignments, containing the designated number of sequences and having the observed width. This product is the expected frequency of observing the particular information content or higher in an arbitrary alignment of random sequences, given the alignment width and the total amount of sequence data. This expectation is called the e-value. The e-value allows the comparison of matrices summarizing differing numbers of sequences and having differing widths. The program can print two different lists of matrices. The first list contains the matrices having the highest information content from each cycle, ordered by decreasing statistical significance (i.e., increasing e-value). In general, this first list will contain the most interesting alignment. The second list contains the matrices saved after the final cycle of the program, also ordered by decreasing statistical significance. In general, this latter list will be useful when the user wishes each sequence to contribute exactly one word to the final alignment (i.e., when the "-n" and "-N" options are not used). In the program's output, the words contained in each matrix are listed in the order of their occurrence in the input sequences. The order is indicated by "integer|integer". The first integer is simply a sequential count of the words, and the second integer indicates during which cycle the word was added to the matrix. The location of a word is indicated by "integer/integer". The first integer indicates which sequence contains the word, and the second integer indicates where in that sequence the word is located. If the first integer is preceded by a minus sign, then the complementary word is the one included in the matrix. The output of the program is sent to the standard output. The input files---those containing the actual sequences and those indicated by the "-f", "-a", and "-i" options---can contain comments according to the following convention. The portion of a line following a ';', '%', or '#' is considered a comment and is ignored. Comments can begin anywhere in a line and always end at the end of the line. The one minor exception is that, to avoid ambiguity, comments in the list of sequences (see the "-f" option below) must be preceded by a blank space when not occurring at the beginning of a line. COMMAND LINE OPTIONS: 0) -h: print these directions. 1) General information -f filename: this file (default: read from the standard input) contains the names of the sequences. The names of the sequences must be less than 512 characters. The corresponding sequence may follow its name if the sequence is enclosed between backslashes (\). Otherwise, the sequence is assumed to be in a separate file having the indicated name. The format of the actual sequences is described at the end of these directions. ADVANCED FEATURES: The following four modifiers can appear in front of the name of the relevant sequence: -c: the sequence is circular. -s integer-integer integer-integer: the positions in the sequence indicated by the integer pairs, inclusive, are seed sequences. If the "-s" modifier is used anywhere in the input file, then the initial set of matrices will only be constructed (i.e., seeded) from the sequences within the marked regions. If this modifier is not used anywhere in the input file, then all the sequences will be used to seed matrices. One or more integer pair can be indicated for a single sequence. However, if no integer pairs are given, the whole sequence will be used for seeding matrices. -i integer-integer integer-integer: the positions in the sequence indicated by the integer pairs, inclusive, are the only positions to be analyzed. -e integer-integer integer-integer: the positions in the sequence indicated by the integer pairs, inclusive, are to be excluded from the analysis. When both the "-i" and "-e" modifiers are used, the intersection of permissible positions is analyzed. When a sequence name is not marked by either the "-i" or "-e" modifier, then the whole sequence is included in the analysis. -L integer: width of the pattern being sought (REQUIRED). -q integer: the maximum number of matrices to save between cycles of the program---i.e., the queue size (default: save 1000 matrices). 2) Alphabet options. The three options in this section are mutually exclusive (default: "-a alphabet"). -a filename: file containing the alphabet and the proportionalities for determining a priori probabilities. Each line contains a letter (a symbol in the alphabet) followed by an optional proportionality (default: 1.0). The proportionality is based on the relative prior probabilities of the letters. For nucleic acids, this might be be the genomic frequency of the bases; however, if the "-d" option is not used, the frequencies observed in your own sequence data are used. In nucleic acid alphabets, a letter and its complement appear on the same line, separated by a colon (a letter can be its own complement, e.g. when using a dimer alphabet). Complementary letters may use the same proportionality. Only the standard 26 letters are permissible; however, when the "-CS" option is used, the alphabet is case sensitive so that a total of 52 different characters are possible. POSSIBLE LINE FORMATS WITHOUT COMPLEMENTARY LETTERS: letter letter proportionality POSSIBLE LINE FORMATS WITH COMPLEMENTARY LETTERS: letter:complement letter:complement proportionality letter:complement proportionality:complement's_proportionality -A alphabet_and_proportionality_information: same as "-a" option, except information appears on the command line (e.g., -A a:t 3 c:g 2). -i filename: (OBSCURE OPTION) same as the "-a" option, except that the symbols of the alphabet are represented by integers rather than by letters. Any integer permitted by the machine is a permissible symbol. Each symbol and its optional complement and proportionality must be on a single line. 3) Alphabet modifiers -d: use the designated prior probabilities of the letters to override the observed frequencies. By default, the program uses the frequencies observed in your own sequence data for the prior probabilities of the letters. However, if the "-d" option is set, the prior probabilities designated in the alphabet information (see section 2 above) are used. If the "-d" option is not set, the "-A", "-a", and "-i" options described in section 2 are still needed for determining the sequence alphabet, but any prior probability information is ignored. -CS: (OBSCURE OPTION) ascii alphabets are case sensitive. option is mutually exclusive with the "-i" option (default: ascii alphabets are case insensitive). This -w: (OBSCURE OPTION) only count letters that are included within the sequence fragments being aligned. When the "-i" or "-e" sequence modifiers are used or the sequences contain forward slashes (/), some sequence data is excluded from the sequences being aligned. This option indicates that only the sequence data being aligned will be counted when determining the observed frequency of each letter. When the "-d" option is not used, this option will influence the determination of the a priori probabilities and, thus, affect the outcome of the alignment. When the "-w" option is not used, all the sequence data will be counted towards determining the observed frequency of each letter. Earlier versions of CONSENSUS (version 6c and earlier) and WCONSENSUS (version 5c and earlier) are equivalent to always having the "-w" option in effect. 4) Options for handling the complement of nucleic acid sequences--the four options in this section are mutually exclusive -c0: ignore the complement (the default option) -c1: include both strands as separate sequences -c2: include both strands as a single sequence (i.e., orientation unknown) -c3: assume pattern is symmetrical 5) Algorithm options the "-pr1" and "-pr2" options are mutually exclusive; the "-l" and "-n" options are mutually exclusive; the "-n" and "-N" options are mutually exclusive; the "-m" option can only be used when the "-n" or "-N" option is used. -pr1: (OBSCURE OPTION) save the top progeny matrices regardless of parentage. -pr2: (OBSCURE OPTION) try to save the top progeny matrices for each parental matrix (the default). This option prevents a strong pattern found in only a subset of the sequences from overwhelming the algorithm and eliminating other potential patterns. This undesirable situation can occur when a subset of the sequences share an evolutionary relationship not common to the majority of the sequences. This option corresponds to the original "consensus" algorithm (Stormo and Hartzell, 1989, PNAS, 86:1183-1187; Hertz et al., 1990, CABIOS, 6:81-92). -l: (lowercase L) seed with the first sequence and proceed linearly through the list. This option results in a significant speed up in the program, but the algorithm becomes dependent on the order of the sequence-file names. This option corresponds to the original "consensus" algorithm (Stormo and Hartzell, 1989, PNAS, 86:1183-1187; Hertz et al., 1990, CABIOS, 6:81-92). -n integer: repeat times and allow per matrix. -N integer: repeat times and allow per matrix. the matrix building cycle a maximum of "integer" each sequence to contribute zero or more words the matrix building cycle a maximum of "integer" each sequence to contribute one or more words -m integer: the minimum distance between the starting points of words within the same matrix pattern; must be a positive integer; can only be used when the "-n" or "-N" option is also used. If the integer is a 1, then there is no restriction on the overlap. If the integer is the same as the integer indicated by the "-L" option, then no overlap is allowed (default: integer indicated by the "-L" option). When the "-c2" option is used (see below), then the "-m" option also indicates the minimum distance between the start of a word and the end of a word on the complementary strand. -t integer: terminate the program "integer" cycles after the current most significant alignment is identified (default: terminate only when the maximum number of matrix building cycles is completed). 6) Output options -pt integer: the number of matrices to print of the top matrices from each cycle (default when NOT using the "-l" option: print 4 matrices; default when using the "-l" option: print no matrices). An integer of -1 means print all the top matrices. -pf integer: the number of matrices to print of the matrices saved from the final cycle (default when NOT using "-n" or "-N" options: print 4 matrices; default when using "-n" or "-N" option: print no matrices). 7) Options indicating how unrecognized symbols are treated (default: u1). Symbols are letters when option "-a" or "-A" is used; symbols are integers when option "-i" is used. The following three options are mutually exclusive. -u0: treat unrecognized symbols as errors and exit the program. -u1: treat unrecognized symbols as discontinuities, but print a warning. Treating a symbol as a discontinuity means that any sequence word containing the unrecognized symbol will be ignored. -u2: treat unrecognized symbols as discontinuities, and print NO warning. FORMAT OF THE SEQUENCE FILES Do not explicitly give the complements of nucleic acid sequences. If needed, the complementary sequence is determined by the program. Whitespace, periods, dashes (unless part of an integer when the "-i" option is used), and comments beginning with ';', '%', or '#' are ignored. When using letter characters (i.e., with the "-a" and "-A" alphabet options), integers are also ignored so that the sequence file can contain positional information. When using integer characters (i.e., with the "-i" alphabet option) the integers must be separated by whitespace. Sequences surrounded by slashes (/) do not contribute to the generation of the patterns; thus, a portion of a sequence can be ignored without disrupting the overall numbering of the sequence. A double slash (//) would indicate a discontinuity in the sequence. A '/' at the beginning or the end of a sequence will cause the sequence to be marked as non-circular even if the sequence's name is marked with a "-c" (see the "-f" option in section 1). The effect of the single slashes can also be created with the "-i" and "-e" modifiers in the file containing the names of the sequences (see the "-f" option in section 1). When slashes and the "-i" and "-e" modifiers are all used, the intersection of permissible positions is analyzed. Sequences that follow their name in the file indicated by the "-f" option must be enclosed between backslashes (\) (i.e., the actual sequence must be preceded and followed by a backslash). However, if the sequence is contained in a separate file, do NOT use a '\'.