Influence of tree shape and species richness on phylogenetic
advertisement

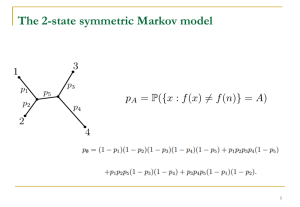
Electronic Supplementary Material 2 ESM 2 - Influence of tree shape and species richness on phylogenetic entropy decomposition In this study, each tree tip was considered as a species. The Yule model assumes a constant speciation/extinction rate along the tree (Yule 1925). Consequently, all branches have the same probability of splitting into two new branches. Trees generated under the Yule model are generally more balanced than observed phylogenies (Mooers & Heard 1997) (see fig. S1a in ESM 2). In the PDA model, every tree with the same number of tips is equally likely (Rosen 1978), but most of the possible arrangements are unbalanced. Conversely, this model generates more unbalanced trees than do empirical phylogenies because the trees are chosen among all possible arrangements (Cunningham 1995) (Fig. S1b in ESM 2). For each combination of tree shape (i.e. more or less balanced) and size (50 and 100 species), we generated 100 replicated trees using the R package “apTreeshape” (Bortolussi et al. 2006). For a given total number of tips, trees had the same total branch length regardless of the model used (i.e. Yule or PDA model). We then sampled a 20-species community from each tree constituting the composition of the first community. To explore the influence of species richness, we built the second community by randomly sampling species along a gradient of richness (from 1 to 50 or 1 to 100 species). We allocated species abundances to each community using a log-normal distribution. Log-normal species-abundance distribution is commonly observed in natural communities, regardless of species richness, spatial scale, or geographic position (Ulrich et al. 2010). The α-diversity of each community was estimated using the phylogenetic entropy of Allen et al. (2009). We then constituted pairs of communities with one of 20-species and the other with n-species (with n ranging from 1 to 50 or 1 to 100) for each of the 100 replicates for each case (shape and richness of the regional tree). Species abundances were summed over the two local communities to calculate the γ-diversity of these two communities. Finally, we quantified the β-diversity between pairs of communities for each of the 100 replicates using phylogenetic entropy decomposition. For a given species richness level, α-, β- (i.e. βst, the proportion of biological turnover) and γ-diversity values were averaged (Fig. 2 in the main text). Relationships among α-, β-, and γ-diversity components calculated on 100-species trees generated under either the Yule or PDA model were investigated using the Spearman coefficient of correlation (Table 2). Communities sampled among 50-species trees were not considered to avoid bias in β-diversity values introduced by the pairs including too many species poor communities. References Allen, B., Kon, M. & Bar-Yam, Y. 2009 A New Phylogenetic Diversity Measure Generalizing the Shannon Index and Its Application to Phyllostomid Bats. Am. Nat. 174, 236-243. (doi: 10.1086/600101) Bortolussi, N., Durand, E., Blum, M. & François, O. 2006 apTreeshape: statistical analysis of phylogenetic tree shape. Bioinformatics, 22, 363-364. (doi: 10.1093/bioinformatics/bti798 ) Cunningham, S. A. 1995 Problems with null models in the study of phylogenetic radiation. Evolution, 49, 1292-1294. Mooers, A. O. & Heard, S. B. 1997 Evolutionary process from phylogenetic tree shape. The Q. Rev. Biol., 72, 31–54. (doi: 10.1086/419657) Rosen, D. E. 1978 Vicariant patterns and historical explanation in biogeography. Syst. Zool., 27, 159–188. (doi:10.2307/2412970 ) Ulrich, W., Ollik, M. & Ugland, K. I. 2010 A meta-analysis of species-abundance distributions. Oikos 119, 1149-1155. (doi: 10.1111/j.1600-0706.2009.18236.x) Yule, G. U. 1925 A mathematical theory of evolution, based on the conclusions of Dr. J. C. Willis, F R. S. Philos. T. R. Soc. Lond., 213, 21-87. Figure S1. Varying tree shapes: (a) a balanced tree generated by the Yule model and (b) an unbalanced tree generated by the PDA model. Both trees the same number of tips (i.e. 50 tips) and total branch length, and belong to our simulated data set. (a) (b)