(spoligotyping) and other bacterial species
advertisement

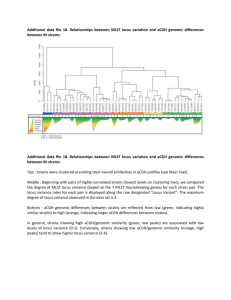
Cluster regularly interspaced short palindromic repeats (CRISPR) are variable loci widely distributed in prokaryotes and are useful for strain typing in Mycobacterium tuberculosis (spoligotyping) and other bacterial species. We tested whether these sequences may be used as natural and specific tags of L. casei strains and how powerful are they as a typing tool for this species. We used the complete genome sequence from two L. casei strains, the type strain ATCC334 and the industrial strain DN-114 001, to identify CRISPR loci and to design flanking primers for their amplification in 20 other L. casei strains of different origins previously characterized by the MLST genotyping method (Diancourt et al, 2007. Appl. Env. Microbiol. 73(20):6601). One CRISPR locus was found in each genome sequence, and these loci were located in a distinct genomic context. When using specific primers flanking the DN114 001 CRISPR locus, PCR amplification was successful in many L. casei strains, whereas amplification with primers flanking the CRISPR locus in strain ATCC 334, which corresponds to a unique genotype based on MLST data, failed to amplify a PCR product in all tested strains. The amplified locus was sequenced in all PCR positive strains, revealing a strong polymorphism of repeat content and sequence. This shows that CRISPR elements in L. casei probably constitute powerful tags for strain-to-strain discrimination. The comparison of L. casei strain relatedness deduced from MLST and CRISPR data will be discussed.