F3 parameters:
advertisement

Introduction:
The purpose of the version 1.0 F3 model is to provide a temporally
and spatially-explicit description of the population dynamics of a coastal
marine fish or invertebrate species. The model allows one to adjust physical
oceanographic parameters describing the current dynamics that disperse
marine larvae, species demographic and population parameters influencing
metapopulation dynamics, and economic rules governing harvesting
strategies (including implementation of marine protected areas). Through
this explicit integration of coastal oceanography, species metapopulation
biology, and fishery harvesting and economics, the F3 model enables one to
examine the interdependent roles of different spatially structured harvesting
methods, post vs. pre-settlement larval density dependence, spiky vs. smooth
subpopulation mixing via larval dispersal, the extent and grain of spatial and
temporal analysis, fixed vs. periodic boundary conditions, and species
demographic parameters on spatial and temporal patterns of fishery harvest
effort and yield, and species escapement from harvest, population
abundance, and larval recruitment.
A.F3 Model
The balance equation describes changes in stocks due to harvest ( H xn ), natural
survivability and larval recruitment.
Axn1 Yxn MYxn Yxn' Fxn' Ln K xn x ' R xn dx'
(1)
where:
Axn 1 adult abundance at location x in the next generation
Yxn Axn H xn escapement at location x (in current generation, n)
= adults at x – the harvest at x (in generation n)
MY xn number of adults (that escaped harvesting) that died due to natural causes
K xn x ' larval dispersal kernel from location x’ to x in generation n
Yxn' number of adults a location x’ (that escaped harvest in generation n)
Fxn' fecundity of adults at location x’
Ln = larval survival (from release through settlement)
Rxn post-settlement recruitment of settled larvae to adults at location x
The terms for fecundity, larval survival and post-settlement recruitment are multiplied
together. Hence, a single constant is used describe fish productivity, Po. Density
dependence is described using the Ricker equation for either fecundity or post-settlement
recruitment.
B. Parameter Descriptions
I. Space/Time Parameters
General Description
This model simulates the growth, reproduction and larval dispersal of a single,
marine species over a given number of generations along a generic coastline of specified
length. The parameters that define aspects of the spatial and temporal aspects of the
model are described in this section.
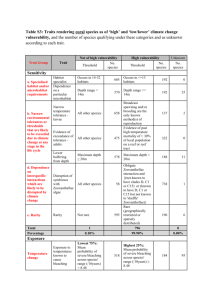
F3Description
The temporal domain of the model simulation is an important aspect of this
model. Ntime sets the number of generations (n) the model will run. It indicates the
number of times the program runs through the Main program loop. The model runs for
each value 1 to Ntime, according to Equation 1. The final simulation would yield,
AxNtime , which represents the adult abundance at location x in the last generation run.
The length of the domain (i.e. coastline) is an important spatial aspect of the
model. The model assumes the species to occupy only a linear habitat along this coast
(e.g. a subtidal reef). The individuals can live along any section of this domain, which is
equal to the parameter rangex [km]. The x-axis in many of the model output figures is
distance from the center of the domain (see Figure 1).
Figure 1. Adult abundance at generation 50 using rangex=400 and delx=5.
The domain is broken up into small sections, the size of which is determined by
the value of delx. For example, a domain with rangex = 400 km and delx = 5 km has a
total of 81 sections or x values. Each value of x is identical in size and is the unit at
which we calculate values such as adult abundance or recruitment. In Figure 1, the
number of adults is calculated for each value of x (i.e. every 5 km), rather than every 1
km along the domain. In Equation 1, x is the specific location we are examining and x’ is
any other location in the domain. If we imagine that the entire domain represents a
metapopulation, then each x could be considered a distinct local population that can
interact with each of the other local populations in the system. This assumes that the
group of individuals within each location x is distinct or somehow isolated from the
group in an adjacent location x.
Another spatial aspect of the domain is what happens at the edges. In the ocean,
there is rarely a finite edge to a particular region of habitat because the system is open
and larval dispersal can be on the scale of hundreds of kilometers. The parameter
periodic defines how the model sets up the edges of the domain. If periodic = 0, then
the model uses fixed, absorbing boundaries at the end of the domain, such that larvae can
disperse out but cannot come back inside. Fixed boundaries are not very realistic because
of the large scale of larval dispersal. If periodic = 1, then the boundaries are periodic,
resulting in a simulated coastline which acts like a series of adjacent domains (i.e. when a
larva reaches the edge of the original domain, it can continue dispersing into an identical/
adjacent domain without altering its dispersal trajectory). Similarly, larvae can travel
from an adjacent domain into the original domain. Therefore it is possible to have inputs
into the domain from outside through the periodic boundary. This will lead to overall
higher adult abundance than with fixed boundaries because there are inputs to the system
that do not occur with fixed boundary conditions. Using periodic boundaries better
reflects reality because a specific location or area in a marine system is connected to
distant locations via larval dispersal, which can occur in spatial scales larger than the
domain size.
II. Mortality and Productivity
General Description
The change in population size is dependent on the fecundity of adults and the
mortality of all stages of individuals. This model incorporates these parameters in several
different ways.
F3 Description
Adult mortality is set by the parameter M, which represents the fraction of
individuals that die each generation due to natural causes (e.g. senescence, predation). In
this model, harvesting mortality occurs first and natural mortality second. Therefore,
according to Equation 1, M impacts only the adults that have not been harvested (i.e. Yxn ,
the escapement).
The productivity of adults in this model is determined by several different
components of Equation 1 (particularly those inside the integral). Po represents the
number of larvae released by each adult (at location x’) per generation that survive
through recruitment. The parameter Po is actually the product of 3 separate biological
parameters from Equation 1:
Fxn' fecundity [# larvae produced / adult at x’],
Ln = larval survival [# larvae produced at x’ that successfully settle
somewhere / total # of larvae produced at x’],
Rxn post-settlement recruitment [# successful recruits / total # successful
settlers]
Multiplying these together gives the number of larvae produced per adult at x’ that
survived through recruitment. Since we don’t have realistic estimates for any of the 3
components, we combine them into a single parameter, Po. Each adult produces large
numbers of larvae yet very few survive dispersal and successfully recruit. Thus, larval
production, natural survival and recruitment success probably multiply together to equal
Po ≈ 1-10. Therefore, in this model, we might use a guess of Po = 2 so that each adult
successfully produces 2 larvae that are capable successfully recruiting. Note that this
number will be further diminished by fracSuccess, which represents the proportion of
larvae that are not “lost” in the ocean due to current advection away from the coast.
Density dependence can impact the fecundity and post-settlement recruitment (see
section IV for a more in depth explanation).
Both M and Po are used to calculate the carrying capacity and the steady state
adult abundance levels (for a uniform environment, with no harvest) (See section A).
III. Harvest
General Description
One of the main goals of this model is to simulate the impact of different types
and levels of harvest on fish abundance and dispersal. A large part of that revolves
around evaluating the effects of marine protected areas (MPAs) along with the different
aspects of harvest. This model allows us to change different parts of all of these human
impacts on the fish population.
F3 Description
The model has two main different harvest parameters: one that sets the fishing
policy (Harvest) and another that determines the intensity of the harvesting (Ho). The
value of the Harvest parameter sets the type of fishing that occurs (i.e. How do the
fishermen go about obtaining the fish? What methods are they utilizing?).
If Harvest = 0, the model uses spatially uniform effort. This means that no
matter how many fish there are in each bin, the fishermen exert the same amount of effort
into each bin. We sometimes refer to this as the “dumb fisherman” strategy because the
fishermen don’t need to know anything about the existing stock to accomplish their task.
For this fishing policy, Ho represents the fraction of the virgin carrying capacity that is
harvested each generation.
If Harvest = 1, the model uses constant catch fishing policy. This means that
there is a Total Allowable Catch (TAC) and the fishermen use their knowledge of the fish
stocks to focus their efforts where fish densities are highest. We refer to this as the “ideal
free fisherman” policy because it allows the fisherman to go where the most fish are and
usually means they don’t overfish where the abundance is already low. For this fishing
policy, Ho is the same: the fraction of the virgin carrying capacity that is harvested each
generation. The TAC is calculated as Ho X virgin carrying capacity X the total size of
the domain.
If Harvest = 2, the model uses a constant escapement fishing policy. This means
that a constant level of escapement at each bin is required for each generation and the
fishermen can vary their effort over the domain as long as the overall escapement ends up
the same. Here, the escapement level = (1-Ho) * virgin carrying capacity [# / km].
Marine Protected Areas (MPAs) in this model can be added to the center of the
domain. SizeMPA sets size of the MPA as a fraction of the length of the domain
(rangex). No fishing is allowed within the MPA. If SizeMPA is set to zero, there is no
MPA in the model.
IV. Pre- and Post-settlement Ricker density dependence
F3 Description:
In the F3 model, the Ricker density dependence function is used to relate adult
fecundity (F) or larval recruitment (R) to adult fish population size (Y, the number of fish
that escape being harvested) at that activity’s location (either x’, the spawning site, or x,
the settlement site). Ultimately, fecundity or recruitment at a location relates to the
number of new recruits in the population at t = n + 1. Thus, the F3 Ricker function is
both temporally and spatially explicit:
n
n n cY x '
x' x'
F F Y e
n
x'
n 1
x
R
F LK
n n
x'
, for pre-settlement density dependence, and
n
n
n cY x
x x' x
Y e
, for post-settlement density dependence.
Pre- and Post-settlement density dependence factors determine whether density
dependence acts upon larval production relative to the number of breeding adults
at location x’, or upon larval settlement relative to the number of adults living at
location x, respectively. For all locations, adult populations sizes are estimated
based on the number of adults present after harvesting occurs (i.e. based on those
adults that escaped being harvested).
“Post = 0” enacts pre-settlement density dependence:
Fxn' Fxn'Yxn' e
cY xn'
,
where c = the Ricker density dependence parameter. Note that Fxn' only equals
the number of larvae that may attempt to disperse. Their dispersal “fate” will
consequentially be influenced by Ln , R xn , fracSuccess, and the Dispersal kernel.
“Post = 1” enacts post-settlement density dependence:
n 1
x
R
F LK
n n
x'
n
n
n cY x
,
x x' x
Y e
In post-settlement density dependence, the function acts upon a larvae population
only after it has successfully survived all the stages prerequisite to recruiting a
location x. Of those larvae that are determined to be ready to settle at each
location x, some are then “culled” by density dependence relative to adult
population size at x.
c: Ricker density dependence function parameter [km/#], affecting the larval
fecundity (pre-settlement), or recruitment (post-) rate’s dependence on adult
population density at that activity’s location. Adjusting c from 0 to 1 changes the
“strength” of the density dependence: At c = 0, larvae fecundity or recruitment
rate is unaffected by density, At c = 1, the rate is affected by exceedingly strong
(ecologically undefendable?) density dependence.
V. The Dispersal Kernel
General Description:
The dispersal kernel is a probability distribution curve meant to approximate the
fluid mechanics of larval transport in a coastal ocean. It is estimated by calculating the
density distribution of larvae settlers based on many individual releases from a single
location. To generate a dispersal kernel through computer simulation, independent
trajectories of thousands of particles, each released from the same coast location, are
simulated in a quasi-realistic velocity field representing ocean flow. Their spatial
patterns are then averaged to estimate the probability that, given a particle lands
somewhere along the coast, it will disperse to a location x distance from its release point.
During simulation, each dispersing particle updates it velocity by receiving a small
random impulse at each time step, resulting in a trajectory similar to a random walk,
except that the steps are serially auto-correlated. Particle impulses are calculated relative
to the mean and random-normal fluctuating velocities of the ocean flow field on a 2dimensional horizontal plane (i.e. ocean flow is assumed to be vertically uniform, or
barotropic). Due to the stochastic turbulence of the flow field, a particle’s serial positions
along its trajectory are expected to be auto-correlated only for a limited time period,
called the Lagrangian decorrelation timescale. The fluctuating velocity of the flow field
is calculated relative to this decorrelation timescale, which increases nonlinearly with
perpendicular distance from the coast, saturating at 3 days at approximately 2 km. Thus,
the motion of a particle more than 2 km from the shore becomes statistically independent
from its motion three days earlier.
Relating particle transport to larval life history, particle ensembles in each
simulation are assigned a precompetency period (time in plankton until larvae are capable
of settling), followed by a competency period (time window during which larvae are
capable of settling). Thus, particle settlement is dependent on being at a coastal location
sometime during its competency period. Aside from these biological factors, larvae are
considered to be purely planktonic particles subject to passive advection.
For each flow field and biological value, averaging over the many individual
independent particle trajectories that successfully disperse enables calculation of a larval
dispersal kernel that sums to 1 (100% probability of dispersing somewhere). Larval
dispersal kernels calculated from simulation processes appear similar to a Gaussian
distribution. Thus, for modeling purposes, a generic dispersal kernel can be described:
K x K o e(
( x xd ) 2
,
2 d2
1
= the amplitude of the probability distribution, xd = the offset, or
( 2 )1 / 2 d
downstream “drift” of the distribution parallel to the coastline relative to a larval release
where K o
location, and σd = the spread of the distribution about the release point. xd = TmU, where
Tm = the planktonic larval duration (PLD, the pre-competency period + the mean of the
competency period), and U = the mean along-shore flow velocity in km/day (e.g. as
caused by the southward-flowing California current). Note: the program asks you to
provide U = the velocity in cm/s, then it converts your input into km/day units. σd =
2.238σuTm½, where σu = the amplitude of the fluctuating flow velocity [km/day], or the
root mean square (RMS) of the velocity ([{v2}mean]½). Note: the program asks you to
provide ustd = the RMS of the velocity in cm/s, then it converts your input into km/day
units.
This dispersal kernel assumes that flow parameters inputted into the Gaussian
function are constant. It also assumes the coastline to be linear, and for variations in
depth and the benthic environment (e.g. reefs) to have no added affect on flow. Specific
to larval dispersal, it assumes larvae to be lagrangian (water-parcel-following).
F3 Description:
The dispersal kernel is used to model transport of larvae from a breeding location
to multiple individual settlement (and, ultimately, recruitment) locations along the
coastline. For a number of larval release events occurring along the coast (ndraw
[days]), and an estimated fraction of those events that will produce larvae that
successfully settle somewhere (fracSuccess), the dispersal kernel will run them through
its probability distribution and determine where each of the larvae from each of the
release locations disperse to along the coast.
The kernel can be parameterized two ways. Choosing MethedofKernelCal =
“function of current” prompts you to provide oceanographic-based estimates of U and
ustd, as well as Tm. The program then uses these estimates to calculate σd and xd for use
in the Gaussian function. Choosing MethedofKernelCal = “fixed kernel” allows you to
directly estimate the mean larval dispersal distance (Dd) and the offset (xd), or
downstream “drift”, larval dispersal parallel to the coastline relative to a release point.
This method is appropriate for those with genetic, microchemistry, or other nonoceanographic estimates of dispersal patterns.
The F3 program can also predict larval dispersal using either a smooth or spiky
kernel. For the smooth kernel (Spiky = 0), the number of larvae produced at a location
during a dispersal event (i.e. an ndraw at a location x’) is fractionalized, then dispersed
along the coastline according to the kernel’s dispersal probability distribution described
above. This generates a smooth dispersal pattern of larvae across the coastline. Setting
Spiky = 1 activates a spatially-discrete version of the smooth kernel. Here, for each
spawning location, x’, a random dispersal trajectory is drawn for each independent
(ndraw) group of dispersing larva from the kernel’s probability distribution. This results
in “spikes” of groups of dispersed larvae to discrete locations along the coastline. Use of
the spiky kernel in this program is recommended because it better emulates real dispersal
patterns. Due to the decorrelation timescale (~3 days), there are a limited number of
independent dispersal events (ndraw) per generation. Furthermore, the probability that a
group of larvae disperse to a coastal location during their competency period
(fracSuccess) is very low. As a consequence, there are only a few larval settlement
events along the entire domain of the coastline per generation, and these events are
clumped spatially. The spiky kernel mimics this clumped dispersal pattern, creating the
high degree of spatial heterogeneity in dispersal pattern that is believed to exist in nature.
Conversely, the smooth kernel does not represent a realistic dispersal pattern occurring
within a single time step (generation), but instead it better describes the long-term
average dispersal pattern for the meta-population.
C. F3 parameters
Space/Time Parameters:
Ntime: sets the number of generations the model will run.
periodic: Designates whether the edges of the domain are fixed or periodic.
periodic = 0: boundaries are fixed, absorbing boundaries.
periodic = 1: boundaries are periodic. At the edge of the domain, another adjacent
domain begins so that larvae can travel in and out of the edge of the domain without
disappearing from the model.
delx: The length of each section (value of x) along the domain [kilometers]
rangex: Sets the size of the domain [kilometers]. For example, if rangex = 400, the
domain would range from –200 to 200 km.
Biological Parameters:
M: The adult mortality rate per generation. In the model, M is the fraction of the
escapement that dies in that generation.
Po: The number of released larvae produced per adult that survive to recruitment. This
parameter is the product of 3 parameters:
Fxn' fecundity [# larvae produced / adult at x’],
Ln = larval survival [# larvae produced at x’ that successfully settle
somewhere / total # of larvae produced at x’],
Rxn post settler recruitment [# successful recruits / total # successful settlers]
Since we don’t have realistic estimates for any of these three parameters, they are
combined into 1 parameter, Po. This parameter describes larval settlement and
recruitment success in terms of biological context only (eg. predation, starvation); it
does not account for settlement failure due to fluid mechanical loss (see
fracSuccess)
Post: Pre- or post-settlement Ricker density dependence
Post = 0: pre-settlement density dependence. For generation t = n, the number of
larvae produced at location x’, as a function of the adult population density at
location x’.
Post = 1: post-settlement density dependence. For generation t = n, the number of
larvae that settle at location x, as a function of the adult population density at
location x. Larvae that successfully settle are considered new recruits that will be
added to the adult population in generation t = n + 1.
c: Ricker density dependence parameter [km/#] affecting the “strength” of the
dependence of larval production (pre-settlement) or settlement (post-) on adult
population density at that activity’s location.
c = 0, no density dependence.
c = 1, excessively strong density dependence.
Harvest Parameters:
Harvest: Sets the fishing policy used in the model.
Harvest = 0, spatially uniform effort (“dumb fisherman” policy)
Harvest = 1, constant catch fishing policy (“ideal free fisherman” policy)
Harvest = 2, constant escapement fishing policy (“constant escapement” policy)
Ho: Determines the intensity of harvest. When Harvest = 0 or 1, Ho is the fraction of
virgin carrying capacity that is harvested each generation. When Harvest = 2, the
escapement level = (1-Ho) * virgin carrying capacity
SizeMPA: Sets the size of the marine protected area as a fraction of the domain size. The
MPA is centered in the domain. If SizeMPA = 0, then there is no MPA.
Kernel Parameters:
Dispersal kernel: A larval settlement probability distribution shaped similar to a
Gaussian curve, such that a larva released at location x = 0 is predicted to have a
probability of dispersing to location x =
Kx = Koe
( x xd ) 2
,
2
2 d
1
= the amplitude of the probability distribution, xd = the drift
( 2 )1 / 2 d
of the ocean current parallel to the coastline (e.g., as caused by the southward-flowing
California current), and σd = the spread of ocean water parallel to the coastline from a
release point. The dispersal kernel is used to distribute larvae along the coastline line.
where K o
Spiky: Sets dispersal kernel to spiky or uniform/smooth.
Spiky = 0: Uses a uniform/smooth kernel with a probability distribution that
integrates to one. For each independent larval release event (i.e. each ndraw at
each x’), the number of larvae that will disperse successfully (calculated via
fracSuccess) is a continuous variable. It is fractionalized, then dispersed along
the coastline according to the smooth kernel’s dispersal probability distribution.
This generates a smooth dispersal pattern of larvae across the coastline.
Spiky = 1: Uses the spiky kernel, which is a spatially-discrete version of the smooth
kernel. For each independent larval release event, a random dispersal trajectory is
drawn for that group of larva from the kernel’s probability distribution. This
results in “spikes” of dispersed individuals to discrete locations along the
coastline.
ndraw: The number of independent larval release events at each location x’ per
generation. Ndraw is approximated by dividing the species’ breeding season [days]
by the ~3-day ocean circulation decorrelation time scale.
fracSuccess: The fraction of ndraw larval release events that successfully return to
anywhere along the coastline during the organism’s settlement competency window.
That is, the proportion of produced larvae that actually disperse and recruit. This
variable limits the number of larvae that “qualify” to pass through the dispersal
kernel.
MethodOfKernalCalc: Determines the method of Kernel calculation.
MethodOfKernalCalc =0, fixed kernel. Choose this option when you have nonoceanographic (e.g. genetic, microchemistry) estimates of the mean dispersal
distance (Dd [km]), and the offset, or downstream “drift”, of the dispersal probability
distribution along the coastline relative to x’ (xd [km]).
MethodOfKernalCalc = 1, function of current. Choose this option when you have
oceanographic estimates of the mean current velocity (U [km/day]), the root mean
square of the current velocity (ustd, [km/day]), and the planktonic larval duration
(Tm [days]).
Dd: Mean larval dispersal distance along the coastline [km]. Applies to the fixed kernel.
When available, use non-oceanographic data to provide an estimate for this parameter.
xd: Offset, or downstream “drift” of the dispersal probability distribution parallel to the
coastline relative to a spawning location [km]. Applies to the fixed kernel. When
available, use non-oceanographic data to provide an estimate for this parameter.
Tm: The planktonic larval duration (PLD). The mean number of days between when a
larva is released and when it settles at its recruitment site. Approximately equal to the
pre-competency period plus the mean of the competency period.
U: Mean along-shore flow velocity [cm/s] parallel to the coastline (e.g., as caused by the
southward-flowing California current). 0.864*U = UU, the along-shore velocity in
km/day. UU*Tm = xd, = the offset, or downstream “drift”, of the Gaussian function
that approximates the dispersal kernel (see Dispersal kernel for further description).
ustd: The amplitude of the fluctuating flow velocity, or the root mean square (RMS) of
the flow velocity [cm/s]. 0.864* ustd = σu [km/day]. 2.238* σu*Tm½ = σd = the
spread of the Gaussian function that approximates the dispersal kernel (see Dispersal
kernel for further description).