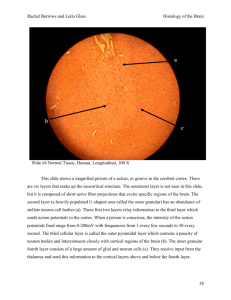
Objetivos: In this work we characterized glial cells and
advertisement

CRUSTACEAN GLIAL CELLS: WHAT ARE THEY LIKE? Silvana Allodi1, Simone Florim da Silva1,2, Clynton Lourenço Corrêa1,3, Cristine Maria Bressan1,4, Ana Maria Blanco Martinez1, Leny Alves Cavalcante5 1 - Programa de Pós-Graduação em Ciências Morfológicas, Instituto de Ciências Biomédicas, Universidade Federal do Rio de Janeiro (UFRJ); 2 – Departamento de Morfologia, Centro de Ciências Médicas, Universidade Federal Fluminense; 3 – Departamento de Fisioterapia, Universidade Federal dos Vales do Jequitinhonha e Mucuri; 4 – Departamento de Biologia Celular, Embriologia e Genética, Universidade Federal de Santa Catarina; 5 – Instituto de Biofísica Carlos Chagas Filho, UFRJ. Objectives: Invertebrates with their more than one million documented species constitute approximately 95 % of all known living organisms on Earth. However many aspects of their nervous system biology, and especially of their glial cells, have not yet been investigated. Therefore, in order to contribute to the knowledge of these cells, in this work we characterized the glial cells of the visual system of two crustaceans: the crab Ucides cordatus and the prawn Macrobrachium rosenbergii. Methods and Results: The eyestalk of decapod crustaceans is constituted by ommatidia and by the lamina ganglionaris (LG), the external (EM), internal (IM) and terminal medullae, which are followed by the protocerebral tract (PT). The LG, made up by monopolar neurons and glia, is the site where the first visual synapses with axons from the ommatidia occur. A thick basement membrane (BM) composed by cells and extracellular matrix separates the ommatidia from the fasciculated zone (FZ), a nerve formed by the ommatidia photoreceptors and glial cells. Classical histochemical reactions for revealing carbohydrates by light microscopy (Periodic Acid-Schiff or PAS), electron microscopy (Thiéry technique), immunohistochemical methods, immunoelectronmicroscopy and Western blot analyses for glial fibrillary acidic protein (GFAP), 2´,3´-cyclic nucleotide 3´-phosphodiesterase (CNPase), glutamine synthetase (GS) and S100 were used. PAS-positive material evidenced glycogen in putative glial cells in different cell processes in specific regions of the optic ganglia of both species, such as the BM, among the photoreceptor axons, surrounding the cartridges and the outer limits of the LG, EM and IM. The Thiéry method revealed glycogen aggregates in different glial cell processes in the FZ and PT, turning visible the presence of at least two types of glial cells in these structures, the perineurial and periaxonal cells. The immunoreactions (for both light and electron microscopy) with anti-GFAP and anti-CNPase, which in vertebrates evidence astrocytes and a non compact myelin protein, respectively, labeled distinct cell types. Immunocytochemical reactions with antibodies against GS full length molecule and S100, as well as the binding of the insect glia and rat astrocytic marker, the Datura stramonium lectin, also evidenced different types of glial cells. Western blotting for GFAP, GS and S100 confirmed their presence in the visual system of crabs and prawns. Conclusions: These results suggest that crustacean glial cells may be studied with classical methods used for a few insects and vertebrates. Our data also indicate that phenotype protein markers of the optic lobe glia share antigenic determinants with GFAP, CNPase, GS and S-100, which have appeared in higher invertebrates and may be conserved in evolution. Financial support: FAPERJ, CNPq, FUJB/UFRJ.