(FXR) predisposes to ileocolonic localization of Crohn`s disease
advertisement

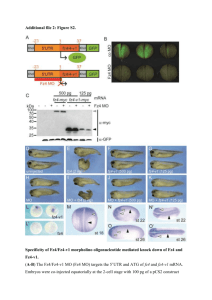
A functional variant of the Farnesoid X Receptor (FXR) predisposes to ileocolonic localization of Crohn’s disease R.M. Nijmeijer1; R.M. Gadaleta2, 3; K.J. van Erpecum2; A.A. van Bodegraven4; J.B.A. Crusius5; G. Dijkstra6; D.W. Hommes7; D.J. De Jong8; C. Ponsioen9; H.W. Verspaget7; R.K. Weersma6; C.J. van der Woude10; M.E. Schipper2; C. Wijmenga11; S.W. van Mil3; B. Oldenburg2 1Department of Surgery, University Medical Center Utrecht, Utrecht; 2Departments of Gastroenterology, Hepatology and Pathology, University Medical Center Utrecht, Utrecht; 3Department of Metabolic and Endocrine Diseases, University Medical Center Utrecht, Utrecht; 4Department of Gastroenterology and Hepatology, VU University Medical Center, Amsterdam; 5Department of Immunogenetics, VU University Medical Center, Amsterdam; 6Department of Gastroenterology and Hepatology, University Medical Center Groningen, Groningen; 7Department of Gastroenterology and Hepatology, Leiden University Medical Center, Leiden; 8Department of Gastroenterology and Hepatology, University Medical Center St. Radboud, Nijmegen; 9Department of Gastroenterology and Hepatology, Academic Medical Center, Amsterdam; 10Department of Gastroenterology and Hepatology, Erasmus Medical Center, Rotterdam; 11Department of Genetics, University Medical Center Groningen and University of Groningen, Groningen. The pathogenesis of IBD is thought to result from a dysregulation of the mucosal immune system and a compromised intestinal epithelial barrier function in genetically predisposed individuals. We recently found that activation of the nuclear bile salt receptor Farnesoid X receptor (FXR) with synthetic FXR agonists improves intestinal barrier function and inflammation in DSS or TNBS murine colitis models. Here we extend these observations to IBD patients. Nine tagging single nucleotide polymorphisms (SNPs) in the FXR gene were selected, 2 of which failed technically. Additionally, two functional SNPs, -1g>t, leading to a defect in translation efficiency, and 518t>c, giving rise to M173T causing reduced FXR function, were added. These SNPs were studied in a Dutch cohort of 1014 Crohn's disease (CD) and 1144 ulcerative colitis (UC) patients, and 934 healthy blood donor controls. To explore a potential relation with disease phenotypes, we analyzed whether FXR polymorphisms were associated with the localization of disease. Furthermore, mRNA expression of FXR and FXR target gene SHP were determined in ileal and cecal biopsies obtained during surveillance colonoscopies in 17 healthy controls and in 14 CD and 12 UC patients in clinical, endoscopic and histological remission. With the current SNP selection, we were able to tag 89% of the FXR gene. No variant of FXR was associated with IBD after correction for multiple testing. Phenotypic information on the localization of the disease was present for 596 patients with Crohn’s disease. The functional variant 518t>c was associated with ileocolonic localization of Crohn’s disease (P=3.54x10-5, OR 5.32, 95%CI 2.2-13.0). Mucosal expression of FXR and SHP were 50% lower in the cecum as compared to the ileum. Ileal FXR expression between controls, CD and UC patients did not differ, but ileal expression of SHP was lower in CD patients compared to controls (p<0.05) and UC patients (p=0.021). A similar trend, although not significant, was observed in the colon. Results were similar when corrected for the villus marker Villin and crypt marker c-Myc. Villin expression was significantly correlated with FXR and SHP expression in healthy controls (R2=0.62, P<0.0002 resp. R2=0.60, P<0.0005). However this correlation was highly reduced in UC patients and completely lost in CD patients. With the present study, we provide additional support for the role of the bile salt nuclear receptor FXR in patients with Crohn’s disease, notably in patients with ileocolonic Crohn’s disease. A therapeutic role of potent synthetic FXR agonists currently available in patients with Crohn colitis needs to be explored.