Restriction Digest of pAMP and pKAN
advertisement
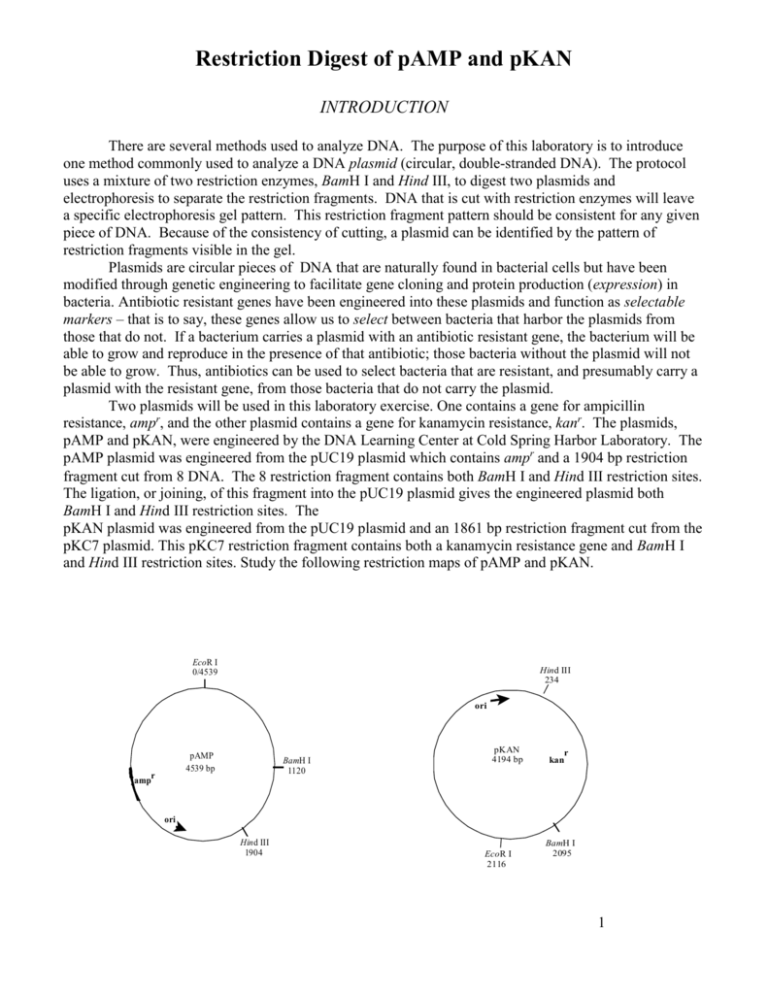
Restriction Digest of pAMP and pKAN INTRODUCTION There are several methods used to analyze DNA. The purpose of this laboratory is to introduce one method commonly used to analyze a DNA plasmid (circular, double-stranded DNA). The protocol uses a mixture of two restriction enzymes, BamH I and Hind III, to digest two plasmids and electrophoresis to separate the restriction fragments. DNA that is cut with restriction enzymes will leave a specific electrophoresis gel pattern. This restriction fragment pattern should be consistent for any given piece of DNA. Because of the consistency of cutting, a plasmid can be identified by the pattern of restriction fragments visible in the gel. Plasmids are circular pieces of DNA that are naturally found in bacterial cells but have been modified through genetic engineering to facilitate gene cloning and protein production (expression) in bacteria. Antibiotic resistant genes have been engineered into these plasmids and function as selectable markers – that is to say, these genes allow us to select between bacteria that harbor the plasmids from those that do not. If a bacterium carries a plasmid with an antibiotic resistant gene, the bacterium will be able to grow and reproduce in the presence of that antibiotic; those bacteria without the plasmid will not be able to grow. Thus, antibiotics can be used to select bacteria that are resistant, and presumably carry a plasmid with the resistant gene, from those bacteria that do not carry the plasmid. Two plasmids will be used in this laboratory exercise. One contains a gene for ampicillin resistance, ampr, and the other plasmid contains a gene for kanamycin resistance, kanr. The plasmids, pAMP and pKAN, were engineered by the DNA Learning Center at Cold Spring Harbor Laboratory. The pAMP plasmid was engineered from the pUC19 plasmid which contains ampr and a 1904 bp restriction fragment cut from DNA. The restriction fragment contains both BamH I and Hind III restriction sites. The ligation, or joining, of this fragment into the pUC19 plasmid gives the engineered plasmid both BamH I and Hind III restriction sites. The pKAN plasmid was engineered from the pUC19 plasmid and an 1861 bp restriction fragment cut from the pKC7 plasmid. This pKC7 restriction fragment contains both a kanamycin resistance gene and BamH I and Hind III restriction sites. Study the following restriction maps of pAMP and pKAN. EcoR I 0/4539 Hind III 234 ori pAMP 4539 bp r amp BamH I 1120 pKAN 4194 bp r kan ori Hind III 1904 EcoR I 2116 BamH I 2095 1 Because Hind III is a specific restriction endonuclease, it will consistently cut DNA wherever it encounters the six-base recognition sequence indicated below. The precise location that is cut is called its restriction site. The DNA molecule consists of two strands of nucleotide building blocks. These building blocks are oriented in the opposite direction on each strand. Thus, the two strands that make-up a DNA molecule are said to be “anti-parallel.” For convenience, we can say that one strand is oriented in a 5’ (“five prime”) to 3’ (“three prime”) direction while the other strand is oriented 3’ to 5’. Careful examination of the restriction sequence will reveal that the sequence of nucleotides is a palindrome; that is to say, it reads the same on both strands when read in a 5’ 3’ direction. 5’…………….AAGCTT……………..3’ 3’…………….TTCGAA……...……...5’ Therefore, whenever Hind III encounters this six-base sequence, it will cut the DNA helix between the adjacent adenine bases. This leaves four unpaired bases forming a “sticky end.” 5’…………A 3’ 3’…………TTCGA 5’ Sticky end 5’ AGCTT…………3’ Sticky end 3’ A………….5’ The recognition sequence for BamH I is: 5’…………….GGATCC……………..3’ 3’…………….CCTAGG……...……...5’ Can you figure out the 5’3 prime direction “sticky ends” for BamH I? MATERIALS Reagents pAMP (60 ng/μL) pKAN (60 ng/μL) Distilled water dH20 Restriction enzyme mix BamH I + Hind III 2.5x restriction buffer Equipment and supplies P-20 micropipette and tips 1.5 mL microfuge tubes Minicentrifuge 37ºC water bath Permanent marker 2 METHODS The plasmids pAMP and pKAN have been cloned in overnight cultures of E. coli (mm294) and purified using a commercial plasmid purification kit. This purification produces a high yield of supercoiled plasmid, which increases the efficiency of transformations. The restriction enzymes, BamH I and Hind III, should always be kept in ice to reduce degradation. Preparing the pAMP and pKAN restriction digest 1. Obtain pAMP, pKAN, 2x buffer and restriction mix from the instructor. Keep these tubes in ice. 2. Use a marker to label four 1.5 mL microfuge tubes as follows: A + = pAMP + BamH I and Hind III A- = uncut pAMP (pAMP without enzymes) K + = pKAN + BamH I and Hind III K - = uncut pKAN (pKAN without enzymes) Include your period and group number on each tube so that you can locate them for the next lab period. 3. The following reaction matrix summarizes the volumes used to set-up the restriction digest. Tube pKAN pAMP 2.5x Buffer dH2O Enzyme Mix Total Volume K+ 4 μL - 4 μL - 2 μL 10 μL K- 4 μL - 4 μL 2 μL - 10 μL A+ - 4 μL 4 μL - 2 μL 10 μL A- - 4 μL 4 μL 2 μL - 10 μL 4. Add 4 μL of pAMP to tubes labeled A+ and A-. Change the tip and add 4 μL of pKAN to tubes labeled K+ and K-. 5. Use a fresh tip and add 4 μL of 2.5x restriction buffer to all tubes. 6. Use a fresh tip and add 2 μL of the enzyme mix, containing BamH I and Hind II, to the tubes labeled A+ and K+ only. 7. Using a fresh tip, add 2 μL of dH2O to the A- and K- tubes. What is the purpose of these tubes? 8. Cap the tubes and use the minicentrifuge to spin down the reagents. 9. Place all four tubes into the 37ºC water bath, and incubate for at least 60 minutes. 10. Following the 60-minute incubation period, the digest can be frozen at -20º C until time is available for electrophoresis 3 Electrophoresis of pAMP and pKAN restriction fragments INTRODUCTION It is important at this stage of our experimental procedure that we need to onfirm that Hind III and BamH I have digested the original plasmids and that we have the correct restriction fragments. Gel electrophoresis is a procedure commonly used to separate fragments of DNA according to molecular size or number of base pairs. DNA fragments will migrate through the agarose maze. DNA, because of the phosphate groups, is negatively charged and will move towards the positive (red) electrode. Because it is easier for small molecules to move through the agarose matrix, they will migrate faster than the larger fragments. Picture a group of cross-country runners that are racing through a dense tropical rain forest. All other factors being equal, the shorter runners will be able to navigate through the tangle of overhanging vines and dense foliage faster than the taller runners. So, smaller DNA fragments will move through the tangle of agarose molecules faster than the longer fragments. We’ll take all of our plasmid samples, digested, and undigested, and use electrophoresis to separate these pieces. You might have predicted that your uncut plasmids would produce only a single DNA band; there’s no reason why you would think otherwise. However, it is likely that two or three bands will appear in the undigested plasmid lanes. This is because plasmids isolated from cells exist in several forms. One form of plasmid is called “supercoiled.” You can visualize this form by thinking of a circular piece of plastic tubing that is twisted. This twisting or supercoiling results in a very compact molecule; one that will move through the gel very quickly for its size. A second plasmid form is called a “nicked-circle” or an “open-circle.” Often a plasmid will experience a break in one of the covalent bonds located in its sugar-phosphate backbone along one of the two nucleotide strands. Repeated freezing and thawing of the plasmid or other rough treatment can cause the break. When this break occurs, the tension stored in the supercoiled plasmid is released as the twisted plasmid unwinds. This circular plasmid form will not move through the agarose gel as easily as the supercoiled form; although it is the same size, in terms of base pairs, it will be located closer to the well than the supercoiled form. The last plasmid form we are likely to see is called the “multimer.” When bacteria replicate plasmids, the plasmids are often replicated so fast that they end up linked together like links in a chain. If two plasmids are linked, the multimer will be twice as large as a single plasmid and will migrate very slowly through the gel. In fact, it will move slower than the nicked-circle. Your uncut pAMP and pKAN samples then may each have three bands that appear in the gel. Starting closest to the well, you might observe a multimer, followed by a nicked-circle band and finally, a fast traveling supercoiled band. We will use a special staining technique that permits us to see the fragments embedded within the gel, then make a photographic record of your gel to document this important step. 4 MATERIALS Reagents pAMP (60 ng/μL) pKAN (60 ng/μL) DNA size marker (1 kb ladder) Agarose (0.8%) 2.5x restriction buffer Distilled water 1x SB buffer 5x loading dye Ethidium Bromide Equipment and supplies P-20 micropipette and tips 1.5 mL microfuge tubes Minicentrifuge Electrophoresis apparatus UV transilluminator Poloroid camera Permanent marker METHODS 1. An 0.8% agarose solution has been prepared and is available in a screw-top culture tube. The agarose has been pre-measured (about 25 mL) for your gel trays. 2. Be sure the sides of the well tray are in the up position before pouring in the agarose solution. When the agarose has solidified, about 20 minutes, carefully remove the comb taking care not to tear the wells. Set the tray into the gel box so that the well-end of the gel is closest to the negative (black) electrode. 3. Fill the box with 1 x SB buffer to a level that just covers the entire surface of the gel to a depth of 1-2 mm. Check to see that the gel is covered with buffer and that no “dimples” appear over the wells. Add more buffer if needed. 4. Determine which samples your team will be loading into the gel: K+ and A+ and ONE of the following: K-, A-, or DNA marker. 5. Add 2uL of loading dye to each of the tubes containing plasmids (2 or 3, depending upon your group – be sure to change tips for each tube. 6. If your group is loading the DNA marker, pick it up from your instructor. This sample already contains the loading dye. 7. Microfuge the tubes. 8. Using a fresh tip for each sample, load 10 uL of each sample into the cells based on the “map” given in class. As you do this, slowly lower the pipette tip below the surface of the buffer directly over, but not into, the well. Putting the tip into the well can damage the wall of the well or puncture the bottom of the well. These are not good things to do! Use two hands to steady the pipettor. Slowly dispense the sample by pushing to the second stop of the pipettor. Because of the loading dye, the sample will have a greater density than the SB buffer. This will allow the sample to sink into the well. 5 SB buffer Important: While holding the button on the second stop, slowly remove the pipette tip from the gel box. If you’ve loaded your sample correctly, the well will be filled with a blue colored solution. 9. Secure the cover to the electrophoresis box and connect the electrical leads to the power supply. Be certain that the anode is connected to the anode (red to red) and the cathode is connected to the cathode (black to black). 10. Turn on the power and set to 130-135 volts. 11. When the loading dye has moved to within 1 cm from the far edge (+end) of the gel, turn off the power supply. Unplug the electrical leads from the power supply by grasping the plugs, not the cords. Remove the cover to the electrophoresis chamber. 12. The instructor will remove the gel and stain the gel, 13. Take a photograph of the gel on the UV light box, and include the photo in your lab write-up. 6









