Annotation of the U. urealyticum Genome
advertisement

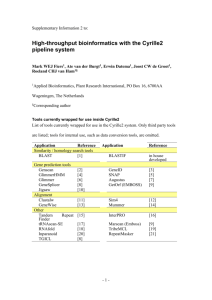
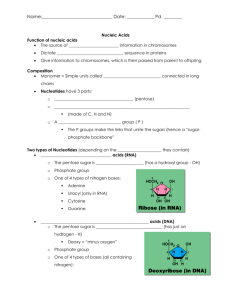
Ureaplasma Genome Paper 02/12/16 Annotation of the U. urealyticum Genome Annotation of the U. urealyticum genome was performed by utilizing a combination of programs for gene prediction, similarity searching, and functional assignment. The information from these analyses was imported into a relational database based upon Microsoft SQL Server. The user interface for this database was a series of web pages that accessed the SQL Server database and allowed us to query available analysis data. Additional pages allowed us to directly hand-annotate the individual gene records thus allowing us to refine start sites and add functional descriptions and notes. This web-based client interface was developed using Microsoft Active Server Pages technology to directly query and update the database records. Basic sequence analysis tools were provided by the Genetics Computer Group package of programs (Wisconsin Package Version 10.0, Genetics Computer Group (GCG), Madison, Wisc.). ORF Identification. Determination of potential protein-coding sequences utilized both Genemark1 and Glimmer2 to create organism-specific open reading frame (ORF) models that could then be used to search the entire genome for ORFs matching the predictive models. The genome was first arranged such that the initial base of the ATG start codon of the putatively-identified dnaA gene was base number 1 of the forward strand. Genemark was run using the predictive model developed from Mycoplasma genitalium to search the U. urealyticum genome for putative ORFs. To ensure that we identified all possible U. urealyticum ORFs, we then utilized Glimmer to create a U. urealyticum-specific model based upon the gene set initially predicted using Genemark, and then derived a second set of putative U. urealyticum ORFs by using Glimmer to search the U. urealyticum gneome a second time with this new model. Each predicted ORF was then assigned a U. urealyticum identification number. ORF UU001 was 1 Ureaplasma Genome Paper 02/12/16 assigned to the putative dnaA gene, and each subsequent ORF was then numbered consecutively according to their left-most base (the start codon for ORFs on the forward strand, and the stop codon for genes on the reverse strand). Subsequent annotation resulted in the identification of a few additional ORFs not originally included in the numbering scheme. These were assigned .1 and .2 designations according to their genomic positions following previously identified ORFs. Gene assignments. BLAST searches were performed on all predicted ORFs using a blastp search of amino acid similarities to sequences in the Genbank non-redundant protein database. The BLAST data was parsed using the blast modules of the BioPerl toolkit (http://www.bioperl.org), and then imported into SQL server tables for analysis. In addition to BLAST similarity searching, we also tentatively identified functional domains within the U. urealyticum ORFs by searching for similarities to the Prosite motif library3, and the Blocks database of protein families4. Programs from the GCG package provided composition and hydrophobicity analysis along with scanning for potential signal peptide domains. The results of these additional analyses allowed us to refine the gene assignments initially made with BLAST. Furthermore, alignments with known proteins provided assistance with start-codon prediction. The results of all of these searches were used to provide putative identification of each U. urealyticum ORF when a significant hit between the U. urealyticum sequence and Genbank sequence was found. A combination of computer-aided gene prediction along with human inspection of each gene record was then used to finalize gene assignments for each U. urealyticum ORF and place each putative gene in one of five categories: Positive assignment. There were published reports that biochemically characterized several ureaplasma genes. 2 Ureaplasma Genome Paper 02/12/16 Putative assignment. There was sufficient sequence similarity with existing genes to suggest functional similarities. Borderline assignment. Sequence similarity with existing genes and/or functional domains existed, but at a borderline significance. Conserved Hypothetical. There was significant similarity with a gene in the database that has been classified as being of unknown function or hypothetical. Unique Hypothetical. There was no significant similarity with any other sequence in the database. RNA identification. To identify genomic sequences that code for tRNAs, the set of programs that encompass the software package, tRNAscan-SE5 was used. Ribosomal RNAs were identified by similarity to the corresponding genes in the Ribosome Database Project sequence database6. The sequences for tmRNA7, the 4.5S signal recognition particle8, and ribonuclease P9 were also identified based upon sequence similarity with known representatives of these RNA genes. Metabolic pathways. Following assignment of coding regions and gene identification, we placed each putatively identified gene into its functional compartment within the metabolic pathways required for the biological functioning of the organism. To facilitate the assignments, we utilized the EcoCyc and MetaCyc packages of metabolic pathway tools10 as a guide. 1. Lukashin, A. V. & Borodovsky, M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26, 1107-1115 (1998). 3 Ureaplasma Genome Paper 02/12/16 2. Salzberg, S. L., Delcher, A. L., Kasif, S. & White, O. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26, 544-548 (1998). 3. Hofmann, K., Bucher, P., Falquet, L. & Bairoch, A. The PROSITE database, its status in 1999. Nucleic Acids Res. 27, 215-219 (1999). 4. Henikoff, S., Henikoff, J. G. & Pietrokovski, S. Blocks+: a non-redundant database of protein alignment blocks derived from multiple compilations. Bioinformatics 15, 471-479 (1999). 5. Lowe, T. M. & Eddy, S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955-964 (1997). 6. Maidak, B. L. et al. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27, 171-173 (1999). 7. Williams, K. P. The tmRNA Website. Nucleic Acids Res. 28, 158-161 (2000). 8. Zwieb, C. & Samuelsson, T. SRPDB (Signal Recognition Particle Database). Nucleic Acids Res. 28, 171-172 (2000). 9. Massire, C., Jaeger, L. & Westhof, E. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol. 279, 773-793 (1998). 10. Karp, P. D. et al. The EcoCyc and MetaCyc databases. Nucleic Acids Res. 28, 56- 59 (2000). 1. Lukashin, A. V. & Borodovsky, M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26, 1107-1115 (1998). 4 Ureaplasma Genome Paper 02/12/16 2. Salzberg, S. L., Delcher, A. L., Kasif, S. & White, O. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26, 544-548 (1998). 3. Hofmann, K., Bucher, P., Falquet, L. & Bairoch, A. The PROSITE database, its status in 1999. Nucleic Acids Res. 27, 215-219 (1999). 4. Henikoff, S., Henikoff, J. G. & Pietrokovski, S. Blocks+: a non-redundant database of protein alignment blocks derived from multiple compilations. Bioinformatics 15, 471-479 (1999). 5. Lowe, T. M. & Eddy, S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955-964 (1997). 6. Maidak, B. L. et al. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27, 171-173 (1999). 7. Williams, K. P. The tmRNA Website. Nucleic Acids Res. 28, 158-161 (2000). 8. Zwieb, C. & Samuelsson, T. SRPDB (Signal Recognition Particle Database). Nucleic Acids Res. 28, 171-172 (2000). 9. Massire, C., Jaeger, L. & Westhof, E. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol. 279, 773-793 (1998). 10. Karp, P. D. et al. The EcoCyc and MetaCyc databases. Nucleic Acids Res. 28, 56- 59 (2000). 5