MS Word file
advertisement

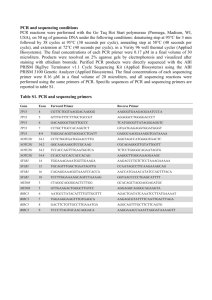
Zainuddin et al SUPPLEMENTARY INFORMATION Methods TP53 mutation detection PCR amplification of exons 4 to 8 of the TP53 gene was performed in the following conditions; 0.2-0.5µg of genomic DNA, 1X PCR buffer, 1.5mM MgCl2, 200µM dNTP, 0.125µM of each primer and 0.7U HotStarTaq DNA polymerase (Qiagen). Final reaction volumes were 50μl each. The cycling program started with an initial cycle of 2-minute denaturation step at 95°C, 1 minute annealing step at 61°C and 1 minute extension step at 72°C, followed by 40 cycles of 30 sec at 95°C, 30 seconds at 61°C and 45 seconds at 72°C. The final extension step was 5 min at 72°C. 7μl of each PCR products was analyzed on a 2% agarose gel in TBE. PCR conditions for p53CA and D17S1678 were similar to the conditions described above. The p53CA PCR reaction mixture was denatured for 3 minutes at 94°C and cycled 35 times (94°C for 1 min, 60°C for 2 min and 72°C for 5 min), followed by a 10-minute extension at 72°C. The D17S1678 PCR reaction mixture cycling program was similar to the p53CA program, except that 56°C annealing temperature was applied. PCR amplification using an alternative set of TP53 primers for each of exon 4 to 8 was performed in the following conditions; 0.2-0.5µg of genomic DNA, 1X PCR buffer, 2mM MgCl2, 200µM dNTP, 0.2µM of each primer and 1U AmpliTaq Gold DNA polymerase (Applied Biosystems). Final reaction volumes were 25μl each. The cycling program started with 40°C for 10 minutes and a 10-minute denaturation step at 95°C, followed by 40 cycles of 30 sec at 94°C, 30 seconds at 56°C and 45 seconds at 72°C. The final extension step was 7 min at 72°C. 7μl of each PCR products was analyzed on a 2% agarose gel in TBE. MDM2 SNP309 and TP53 codon 72 genotyping PCR amplification was made in the following conditions; 0.2-0.5µg of genomic DNA, 1X PCR buffer, 1.5mM MgCl2, 800µM dNTP, 2µM of each primer and 1U HotStarTaq DNA polymerase (Qiagen). Final reaction volumes were 50μl each. The cycling program started with an initial cycle of 10-minute denaturation step at 95°C, followed by 35 cycles of 30 sec at 95°C, 30 seconds at 57°C and 1 minute at 72°C. The final extension step was 7 min at 72°C. To differ between MDM2 genotypes, the resulting 154bp PCR products were digested with 5 units of MspA1 I (New England Biolabs, Beverly, MA) at 37°C for 16 hours. The 1 Zainuddin et al digested products were then electrophoresed on a polyacrylamide gel using Genegel™ Excel 12.5/24 kit (GE Healthcare Bio-Sciences AB) at 14°C for 1 hour and 40 minutes and silverstained. The G/G homozygote product is cleaved by MspA1 I and yields 59- and 95bp bands, the T/T homozygote is not cleaved by the enzyme and yields a single 154bp band, whereas the T/G heterozygote contains all three bands. The 312bp PCR products of TP53 exon 4 were digested with 1 unit of BstUI (New England Biolabs, Beverly, MA) at 60°C for 5 hours. The codon 72 WT Arg/Arg homozygote product is cleaved by BstUI and yields 259- and 53bp bands, the Pro/Pro homozygote is not cleaved by the enzyme and yields a single 312bp band, while the Arg/Pro heterozygote contains all three bands. 2 Zainuddin et al Supplementary Table 1. Primers for amplification of TP53 mutationa and MDM2 SNP309 analysis. Primer name Primer sequence (5’3’) TP53 exons 4 to 8b (GenBank accession number X54156) ACTGAAGACCCAGGTCCAGATGAA p53 – 4 sense AGGGTGAAGAGGAATCCCAAAGTT p53 – 4 antisense TGTTCACTTGTGCCCTGACTTTCA p53 – 5-6 sense p53 – 5-6 antisense AGGGCCACTGACAACCACCCTTA CAAGGCGCACTGGCCTCATCTT p53 – 7-8 sense p53 – 7-8 antisense ACCGCTTCTTGTCCTGCTTGCTTA Alternative set of TP53 exons 4 to 8 primersc CCTGGTCCTCTGACTGCTCT p53 – 4 sense GCCAGGCATTGAAGTCTCAT p53 – 4 antisense TGTTCACTTGTGCCCTGACT p53 – 5 sense AACCAGCCCTGTCGTCTCT p53 – 5 antisense CAGGCCTCTGATTCCTCACT p53 – 6 sense CTTAACCCCTCCTCCCAGCAGAG p53 – 6 antisense TCATCTTGGGCCTGTGTTATC p53 – 7 sense GGGTCAGAGGCAAGCAGA p53 – 7 antisense GGGACAGGTAGGACCTGATTT p53 – 8 sense AGGAAAGAGGCAAGGAAAGG p53 – 8 antisense Fragment size (bp) 312 487 611 362 271 185 203 287 Genomic location TP53 exon 4 TP53 intron 4 TP53 intron 4 TP53 intron 6 TP53 intron 6 TP53 intron 8 including splice site TP53 intron 3 TP53 intron 4 TP53 intron 4 TP53 intron 5 TP53 intron 5 TP53 intron 6 TP53 intron 6 TP53 intron 7 TP53 intron 7 TP53 intron 8 17p deletion (p53CA; GenBank accession number X61505 and D17S1678; GenBank accession number G06085) AGGGATACTATTCAGCCCGAGGTG p53CA-GT 135 25kb upstream from TP53 ACTGCCACTCCTTGCCCCATTC p53CA-AC TTTGGGTCTTTGAACCCTTG WI-9178-1 116 40kb telomeric to CCACAACAAAACACCAGTGC WI-9178-2 TP53 MDM SNP309d (GenBank accession number AF527840) CTGCCCACTGAACCGGC MDM2 sense GAGGTCTCCGCGGGAGTTC MDM2 antisense 154 First intron of the MDM2 promoter a TP53 codon 72 polymorphism was assessed using the PCR products of TP53 exon 4. Montesinos-Rongen M, Roers A, Kuppers R, Rajewsky K, Hansmann ML. Mutation of the p53 gene is not a typical feature of Hodgkin and Reed-Sternberg cells in Hodgkins disease. Blood 1999; 94: 1755-60. cCases which showed no PCR product using the standard PCR primer set were evaluated using an alternative set of TP53 primers for each of exon 4 to 8. d Bougeard G, Baert-Desurmont S, Tournier I, Vasseur S, Martin C, Brugieres L, et al. Impact of the MDM2 SNP309 and p53 Arg72Pro polymorphism on age of tumour onset in LiFraumeni syndrome. J Med Genet 2006;43 (6):531-3. b 3