pbi12424-sup-0017-TextS2
advertisement

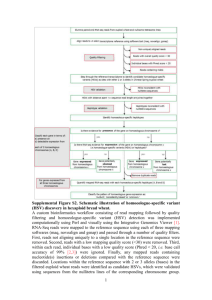
Text S2_Supporting Information Co-location and co-linearity analysis of NCCR heading date QTLs A co-location analysis of QTL supporting intervals (QTL CIs) in the NCCR map with previously reported QTL and meta-QTL for heading date (HD) and a wheat-grasses gene-content co-linearity analysis in the same QTL CIs was carried out and is reported here. The analysis takes advantage of the availability within NCCR of QTL map information based on SNP developed from transcribed genes. Such type of SNPs are more informative in terms of synteny analysis (as extensively reported in Maccaferri et al. 2014). The QTLs were QHd.ubo-2A, QHd.ubo-2B and QHd.ubo-7A.2. The same loci appear also to influence maturity date, as highlighted by the detection of QMd.ubo-2A.1, QMd.ubo2B.1 and QMd.ubo-7A.2 QTLs (See Table 4 in main text). Besides the relevance of these QTLs for the determinism of phenology, we considered the QTLs as case studies to demonstrate that the CI resolution of QTL analysis and the use of transcript-associated SNPs, coupled with barleyBrachypodium-rice co-linearity analysis, allowed us to define the gene space harboring the QTL causal genes to intervals containing less than one hundred genes in all three cases. Comparisons with previously reported HD QTLs approximately co-mapping with PPD and FT loci in hexaploid and tetraploid wheat was difficult because of differences in marker systems (i.e., SNPs vs simple sequence repeats and hybridization-based markers derived from genomic clones). Therefore, the comparison of confidence intervals across maps was carried out based on normalized genetic distances, relative to the total chromosome length, assuming nearly complete genome coverage in both the tetraploid NCCR SNP-based map and the SSR/DArT hexaploid wheat consensus maps from previously published QTL meta-analyses, such as the SSR consensus map Ta-SSR-2004 (Somers et al. 2004), and the map used in the meta-QTL analysis reported by Griffiths et al. 2009). CI boundaries of major PPD- and FT- QTL reported from other maps were first projected on the Ta-SSR2004. In NCCR, the normalized CIs for QHd.ubo-2A and QHd.ubo-2B were comparable to each other (relative positions of 19.9-23.7 and 14.9-20.3 for IBD-CIM, 3.5-24.7 and 14.9-21.7 for IBS-SMA). The meta-QTL 1 for HD reported by Griffiths et al. 2009 in chr 2A and corresponding to PPD-A1 was mapped to a 7.1-32.8 relative position CI. In durum wheat Kofa x Svevo, QHd-2A.2 (Maccaferri et al. 2008) corresponding to PPD-A1 was mapped to a 16.9-22.8 relative position CI (projected on Ta-SSR2004). In chr 2B, the QTL for HD mapped by Bennett et al. (2012), corresponding to PPD-B1, was mapped to a 19.1-27.1 relative interval. The meta-QTL reported by Hanocq et al. (2007) and the QTL in durum wheat Kofa x Svevo, chromosome 2B, both centered to the SSR marker gwm148, were mapped to an interval of 30.0-44.5%. These intervals were very close to the relative intervals of NCCR QTL. In NCCR, QHd.ubo-7A.2 was mapped to the relative interval of 20.8-34.5 (the IBS-SMA included the IBD-CIM interval). This interval is compatible to those reported in Bennett et al. (2012), equal to 34.7-41.5, in Hanocq et al. (2007), equal to 36.8-51.1, and in Griffitths et al. (2009), with 7A meta-QTL1 located to an interval of 9.0-19.0 and 7A meta-QTL2 to 33.5-49.03. Bonnin et al. (2008) mapped TaFT in Courtot x Chinese Spring very close to barc154, whose relative position in the TaSSR-2004 map is 34.5. Since PPD-1 and TaFT have been cloned (Turner et al. 2005; Yan et al. 2006) the gene-based SNP feature of the NCCR map was exploited to proof the potential of the map for mapping QTL down to the candidate gene level. PPD-1 locus was found to contain Ta-PRR73-A1 in wheat, HvPRR37 in barley, and Bradi1g16490 and Os07g49460 as horthologs in Brachypodium and rice (reviewed by Higgins et al. 2010). TaFT locus (VRN3 in wheat) was shown to be horthologous of HvFT1-7H in barley, OsFTL2-Hd3a = Os06g06320 in rice and of Bradi1g48830 in Brachypodium. The genes tagged by mapped SNPs within the CI of the three QTLs (32, 54 and 26 SNP, respectively) were subjected to a search for horthologs in barley, Brachypodium and rice in order to define the main conserved syntenic blocks among grasses in the QTL regions. As a first step, the ca. 200 bases of the Illumina SNP assay from the wheat 90K Illumina assay manifest file were associated to unique Triticum turgidum transcript contigs using BLASTN and the database stored in (http://wheat.pw.usda.gov/GG2/WheatTranscriptome/, Krasileva et al. 2013). The Triticum turgidum transcript contigs were used to retrieve the corresponding orthologs in barley, Brachypodium and rice genomes using the bidirectional best hit approach implemented in ChromoWIZ software, a tool for co-linearity analysis in grass genomes (http://pgsb.helmholtz-muenchen.de/cgibin/db2/chromowiz, Nussbaumer et al. 2014). The results are reported in File S2. In case of QHd.ubo-2A, starting from 16 unique Triticum turgidum transcripts for which at least one grass orthologs were identified among barley, Brachypodium and rice (File S2), the number of orthologs retrieved with the BBH method were 12, 15 and 11 for barley, Brachypodium and rice, respectively. The IBD-CIM CI pointed out a strong conservation of co-linearity between wheat and barley, with a unique barley genomic bin of 10 Mb size in chromosome 2H (the third from the top of chromosome), containing 76 genes, and a more fragmented correspondence with Brachypodium and rice, with three main distinct syntenic blocks in Brachypodium (chromosome 1, 3 and 5), and rice chromosome 7 and chromosome 2. Besides of the unique clearly defined barley bin, the PPD-1 causal gene was clearly confined within a six SNP array including the syntenic block in Brachypodium from Bradi1g16470 to Bradi1g16590 and in rice (from Os07g49480 to Os07g49300) that spanned the causal locus (Bradi1g16490 = Os07g49460). The synteny relationships were less defined for QHd.ubo-2B, which mapped in a chromosome position orthologous to QHd.ubo-2A. In the case of QHd.ubo-2B, the CI spanned a wider interval (14.08 cM) with a lower density of mapped SNPs as compared to the corresponding chromosome region in chromosome 2A. This was probably a consequence of the less defined founder SNP molecular haplotype patterns in the region. Based on the six transcript mapped in the QHd.ubo-2B CI, four barley genomic bins were tagged, of which two in chromosome 2H (including the bin identified for QHd.ubo-2A), one in chromosome 1H and one in chromosome 5H, for a total of 260 genes. The region was therefore much wider than that pointed out for QHd.ubo-2A. Moreover, the QTL peak and CI identified based on IBD-CIM and IBS-SIM were not coincident, with the CI for IBD-CIM more accurate than that resulted from IBS-SIM, based on the chromosome interval harboring the causal gene Bradi1g16490 (File S2). The QHd.ubo-7A.2 CI included 8 transcript that clearly identified barley bin fifth in chromosome 7H including 62 genes, among which the causal FT gene (Bradi1g48830 = Os06g06320). The candidate region was also clearly identified by gene the co-linearity in Brachypodium and in rice. REFERENCES Bennett, D., Izanloo, A., Reynolds, M., Kuchel, H., Langridge, P. and Schnurbusch, T. (2012) Genetic dissection of grain yield and physical grain quality in bread wheat (Triticum aestium L.) under water-limited environments. Theoretical and Applied Genetics, 125, 255-271. Bonnin, I., Rousset, M., Madur, D., Sourdille, P., Dupuits, C., Brunel, D. and Goldringer, I. (2008) FT genome A and D polymorphisms are associated with the variation of earliness components in hexaploid wheat. Theoretical and Applied Genetics, 116, 383-394. Griffiths, S., Simmonds, J., Leverington, M., Wang, Y., Fish, L., Sayers, L., Alibert, L., Orford, S., Wingen, L., Herry, L., Faure, S., Laurie, D., Bilham, L. and Snape, J. (2009) Meta-QTL analysis of the genetic control of ear emergence in elite European winter wheat germplasm. Theoretical and Applied Genetics, 119, 383-395. Hanocq, E., Laperche, A., Jaminon, O., Lainé, A.L. and Le Gouis, J. (2007) Most significant genome regions involved in the control of earliness traits in bread wheat, as revealed by QTL meta-analysis. Theoretical and Applied Genetics, 114, 569-584. Higgins, J.A., Bailey, P.C. and Laurie, D.A. (2010) Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS One, 5, e10065. Maccaferri, M., Sanguineti, M.C., Corneti, S., Araus Ortega, J.L., Ben Salem, M., Bort, J., DeAmbrogio, E., del Moral, L.F.G., Demontis, A., El-Ahmed, A., Maalouf, F., Machlab, H., Martos, V., Moragues, M., Motawaj, J., Nachit, M., Nserallah, N., Ouabbou, H., Royo, C., Slama, A. and Tuberosa, R. (2008) Quantitative trait loci for grain yield and adaptation of durum wheat (Triticum durum Desf.) across a wide range of water availability. Genetics, 178, 489-511. Maccaferri, M., Ricci, A., Salvi, S., Milner, S.G., Noli, E., Martelli, P., Casadio, R., Akhunov, E., Scalabrin, S., Vendramin, V., Ammar, K., Blanco, A., Desiderio, F., Distelfeld, A., Dubcovsky, J., Fahima, T., Faris, J., Korol, A., Massi, A., Mastrangelo, A.M., Morgante, M., Pozniak, C., N’Diaye, A., Xu, S. and Tuberosa, R. (2014) A high-density, SNP-based consensus map of tetraploid wheat as a bridge to integrate durum and bread wheat genomics and breeding. Plant Biotechnology Journal, doi: 10.1111/pbi.12288. Somers, D.J., Isaac, P. and Edwards, K. (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theoretical and Applied Genetics, 109, 1105-1114. Turner, A., Beales, J., Faure, S., Dunford, R.P. and Laurie, D.A. (2005) The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science, 310, 1031-1034. Yan, L., Fu, D., Li, C., Blechl, A., Tranquilli, G., Bonafede, :M., Sanchez, A., Valarik, M., Yasuda, S. and Dubcovsky, J. (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proceedings of the National Academy of Sciences U S A, 103, 19581-19586.