Affymetrix Labeling Protocols: Day1 First Strand Synthesis: Add 5 ug
advertisement

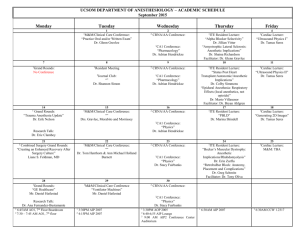
Affymetrix Labeling Protocols: Day1 First Strand Synthesis: 1. Add 5 ug total RNA Primer DEPC water x ul 1 ul y ul 10 ul 2. Incubate at 70C for 10 min. Spin and put on ice. 3. Add 5X first strand buffer 0.1M DTT 2 ul 10 mM dNTP mix 1 ul 4 ul 4. Incubate at 42C for 2 min. 5. Add Superscript II (200 U/ul from kit) 1ul/5ug total RNA or 1 ug mRNA. Total volume will be 20 ul 6. Incubate 1 hour at 42C. 7. Place on ice. Second Strand Synthesis: 1. Add DEPC water 5X second strand buffer 10mM dACTGs E. coli 10 U/ul DNA ligase E. coli 10 U/ul DNA polymerase I E. coli 2 U/ul RNase H 91 ul 30 ul 3 ul 1 ul 4 ul 1 ul 2. Mix gently, centrifuge and incubate at 16C (Reagent Shelf) 2 hours. 3. Add 2ul (10 U) T4 DNA polymerase 4. Incubate at 16C for 5 min. 5. Add 10 ul 0.5 M EDTA (Reagent Shelf) 6. Store at –20C or continue to cleanup. Cleanup of Double Stranded DNA: 1. Pellet Phase-Loc gel at 12,000x g for 30 sec. 2. Add 162 ul (equal volume) of Phenol Chloroform Isoamyl (25:24:1 pH 7.9/8.0) to final volume of 324 ul. 3. Vortex and transfer to PLG tubes. 4. Do Not Vortex again. Centrifuge at 12,000x g for 2 min. 5. Transfer aqueous upper phase to 1.5 ml tube. 6. Add 0.5 vol. 7.5 M Ammonium Ac + 2.5 vol. 100% ethanol (-20) to sample and vortex. (optional: add 1 ul (5mg/ml) glycogen as carrier). Precipitate overnight at –20C. Affymetrix Labeling Protocols Day 2 cDNA Cleanup (continued) 7. Centrifuge at 12,000x g for 20 min. at room temp. 8. Remove supernatant. Wash with 80% ethanol (-20). 9. Centrifuge at 12,000x g for 5 min. 10. Remove ethanol and repeat steps 8 & 9 once more. 11. Air dry pellet and resuspend in 12 ul DEPC water. 12. Store at –20C or continue. RNA Labeling (IVT) 1. Determine volume of sample to give ~1 ug cDNA for template** check with Affy: Original total RNA (ug) resuspension vol 5.0-8.0 8.1-16.0 16.1-24.0 24.1-32.0 32.1-40.0 Volume cDNA for IVT **based on 12 ul 10 ul 5 ul 3.3 ul 2.5 ul 2 ul 2. Add in order to new microfuge tube: Template cDNA x ul DEPC water y ul (22 – x) 10X HY Reaction Buffer (Vial 1) 4 ul 10X Biotin labeled Ribonucleases (Vial 2) 4 ul 10X DTT (Vial 3) 4 ul 10X RNase Inhibitor Mix (Vial 4) 4 ul 20X T7 RNA Polymerase (Vial 5) 2 ul 40 ul total volume 3. Mix reagents carefully and spin briefly. 4. Incubate at 37C for 4-5 hours, mixing every 30-45 min. 5. Clean-up immediately or store at -20C or -70C Cleaning Up IVT Products 1. Purify only one-half of IVT product initially (20 ul), then check yield. Save aliquot of unpurified product for gel electrophoresis before purifying other half. 2. Add 80 ul RNase-free water to 20 ul of IVT product for final volume of 100 ul. 3. Add 350 ul Buffer RLT and mix thoroughly. 4. Add 250 ul ethanol (96-100%) and mix well by pipetting. Do not centrifuge. 5. Apply sample to an RNeasy mini spin column in a collection tube. Centrifuge for 15 sec at ?8000 g. 6. Return flow-through to spin column and centrifuge again for 15 sec at ?8000 g. Discard second flow-through (not compatible with bleach). 7. Transfer spin column to new 2 ml collection tube. Add 500 ul Buffer RPE and centrifuge for 15 sec at ?8000 g. Discard flow-through and reuse collection tube. 8. Add 500 ul Buffer RPE to spin column and centrifuge for 2 min at max speed to dry membrane. Optional: Place spin column in new collection tube and centrifuge at max speed for 1 min to eliminate any possible carryover of Buffer RPE. 9. Transfer spin column to 1.5 ml collection tube and pipette 30-50 ul of RNase-free water (warmed) onto spin column membrane. Wait 1 min and centrifuge for 1 min at ?8000 g to elute cRNA. Repeat if expected cRNA yield is >30 ug (expected yield could be 50 ug from 0.5 ug cDNA). 10. Quantify cRNA concentration and purity before purifying second half of IVT product. Quantifying the Purified cRNA 1. Check OD at 260 nm and 280 nm. Need A260/A280 ratio between 1.9-2.1 2.0 for acceptable purity. 2. Calculate cRNA yield: cRNA (ug) = A260 * 40 ug/ml * [spec vol (ul) / IVT product in spec (ul)] * elution vol (ml) 3. Calculate adjusted cRNA yield if started with total RNA: Adjusted cRNA yield = RNAm – (total RNAi)(y) RNAm = amount of cRNA measured after IVT (ug) total RNAi = starting amount of total RNA (ug) y = fraction of cDNA reaction used in IVT 4. Determine overall concentration of adjusted cRNA product from both halves of purified IVT produced. If cRNA concentration is < 0.6 ug/ul, precipitate sample and resuspend in appropriate volume of RNase-free water. Ethanol Precipitation 1. Add 0.5 volumes NH4Ac to purified cRNA sample. 2. Add 2.5 volumes (sample vol + NH4Ac) of absolute ethanol (-20C) and vortex. Optional: add 1 ul glycogen to help precipitate and visualize cRNA. 3. Precipitate at -20C overnight. Affymetrix Labeling Protocols Day 3 Ethanol Precipitation (cont.) 1. Spin precipitated sample at ?12,000 g at 4C for 30 min. 2. Wash pellet twice with 500 ul of 80% ethanol (-20C). Air dry pellet before resuspension. 3. Resuspend dried pellet in appropriate volume of RNase-free water. Final concentration (adjusted) of cRNA should be at least 0.6 ug/ul. Fragmenting the cRNA 1. Determine (adjusted) amount of fragmented cRNA (x) needed for gel analysis (at least 1 ug) plus hybridization cocktail: Array type Micro/Mini Midi Standard Fragmented cRNA (for one probe array) 5 ug 10 ug 15 ug 2. Calculate amount of RNase-free water and 5X Fragmentation buffer needed to have a final cRNA concentration between 0.5 ug/ul and 2 ug/ul: a. Determine final volume (z) of the fragmentation reaction: x ug cRNA to x ug cRNA 2 ug/ul 0.5 ug/ul b. 5X Fragmentation buffer should be 20% of final volume (2 ul per 8 ul RNA + water) c. Final Fragmentation x cRNA 5X Frag. buffer RNase-free water FINAL VOLUME Reaction: a ul (set by conc. of purified IVT product) b ul (0.2z) c ul (z – a – b) z ul 3. Incubate fragmentation reaction at 94C for 35 min. 4. Chill on ice, then spin to collect condensation. 5. Save aliquot (at least 1 ug RNA) for gel electrophoresis. Store rest of sample at –20C until ready to perform hybridization. Gel Electrophoresis 1. Run aliquots of cDNA (optional), unpurified and purified cRNA, and fragmented cRNA on a 1% agarose gel (or denaturing gel) to estimate yield and size distribution of labeled transcripts. 2. Put 0.3 g agarose in 30 ml 1X TBE and heat until boiling. 3. Cool on shaker table until touchable, then add 1 ul ethidium bromide. 4. Pour gel and let sit until hard. 5. Mix RNA (samples or ladder) with 6X loading dye and heat to 65C for 5 min before loading. 6. Run at 105 V for 30 – 60 min. 7. Fragmented cRNA should be distributed between 35 and 200 bp. Comparing unpurified to purified IVT product can help determine the amount of loss during clean up, if similar amounts or proportions of cRNA are loaded. Preparing Hybridization Target 1. Mix according to Table 5-1 in Affy manual: Fragmented cRNA Control B2 (Thaw 65C for 5 min) 20X Hybridization Controls (Thaw 65C for 5 min) Herring Sperm BSA 2X Hybridization Buffer Water 2. Equilibrate probe array to room temp. 3. Heat Hyb. cocktail to 99C 5 min. in heat block 4. Fill array with 1X hyb buffer and incubate at 45C in oven with rotation for 10 min. 5. Transfer hyb cocktail to oven for 5 min at 45C. 6. Spin hyb cocktail at max. speed in centrifuge 5 min. 7. Remove buffer from array and replace with appropriate volume of clarified hyb cocktail. Avoid any solid material on bottom of tube. 8. Hybridize 16 hours at 45C and 60rpm. Affymetrix Protocol Day 4 End of Hybridization 9. Remove hyb. cocktail. Store on ice or in -20 for long-term storage. Fill probe array completely with non-stringent buffer. Probe array can now be stored for 3 hours at 4C. Preparation of SAPE (Streptavidin Phycoerythrin) and Antibody (if required) SAPE (single stain) for Arrays with 50 um spots For each array: 2x MES Buffer (stored at 4C) 50 mg/ml BSA (Affy box -20) 1 mg/ml SAPE (stored at 4C) DI water TOTAL 300 ul 24 ul 6 ul 270 ul 600 ul SAPE (Antibody Amplification) for arrays with 24 um spots For each array: 2X MES stain buffer 50 mg/ml BSA 1 mg/ml SAPE DI water TOTAL 600 ul 48 ul 12 ul 540 ul 1200 ul Mix and divide into 2 aliquots of 600 ul each. These will be used for stain 1 and 3. Store in foil on ice. Antibody Solution For each array: 2X MES stain buffer 50 mg/ml BSA 10 mg/ml normal goat IgG 300 ul 24 ul 6 ul 0.5 mg/ml biotinylated antibody DI water TOTAL 3.6 ul 266.4 ul 600 ul Run Fluidics Station for Staining and Washing Scan Chip and Analyze Doc Word.Document.8ô9²q