ESM 6 Methods MSVAR uses simulation to infer the coalescent (and
advertisement

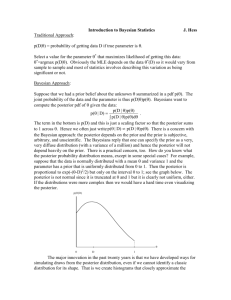
ESM 6 Methods MSVAR uses simulation to infer the coalescent (and thus demographic) process leading to the observed microsatellite allele frequencies. The program assumes a deterministic model of change in effective population size. We chose an exponential growth model, as it seems to better reflect the biology of T. spinipes: Queen are expected to live up to 7-8 years, colonies are believed to produce more than one swarm per year, and the mortality of new colonies seems to be low. MSVAR uses Markov chain Monte Carlo (MCMC) to draw samples from the joint posterior distribution of current and ancestral effective population size, as well as time since expansion or decline. All microsatellite loci are considered simultaneously, as MSVAR uses a hierarchical model where the parameters for each loci are assumed to follow from a (base 10) log-normal distribution parameterized by log mean and dispersion. These population-level (across loci) parameters are current effective population size, ancestral effective population size, mutation rate, and time since population expansion/decline. We use prior modes of 10 4, 104, 10-3.5, and 104; and prior variances of 2, 2, and 0.5 (on the log-10 scale) respectively. For the variances of these parameters across loci, we assume log-normal priors with means 0 and variances 0.5, 0.5, 0.25, and 0.5 respectively. To fit the models, we ran three Markov chains for 500 million iterations each, retaining a sample every 50k iterations for a total of 10k samples from the posterior. We discard the first 5k samples from each thinned chain, and then assessed convergence visually and with the Gelman-Rubin scale-reduction factor (Rhat, Gelman and Rubin 1992). We summarized the marginal posterior distributions of parameters by their expectations and 95% credibility intervals (based on quantiles of the posterior). We estimated the probability that the population experienced a decline (rather than an expansion) as the proportion of MCMC samples where the current effective population size is lower than the ancestral population size. Finally, to assess sensitivity to the choice of priors, we ran the model 8 times, using all combinations of values 10 2 and 104 for the prior modes of current effective population size, ancestral population, and time since expansion/decline. Results The results of the microsatellite coalescent simulations in MSVAR indicate that Trigona spinipes experienced a population decline rather than a population expansion (the estimated posterior probability of decline was 0.95). The current and ancestral effective population size were estimated at 103.52 (95% CI 102.52 to 104.62) and 104.43 (95% CI 103.94 to 104.94) respectively (Figure ESM 6A-A, Table ESM 6B). The posterior expectation of time since decline was 10 2.83 (95% CI 101.97 to 103.80, Figure ESM 6A-B). We found similar results across different choices of priors (Table ESM 6C), as all of the priors considered indicate a population decline occurring between 100 and 1000 years in the past. Due to a correlation between the posterior of current effective population size and the time since decline, the current effective population size was influenced by the prior (dropping from ~10 3.4 under a prior of historic decline to ~102.6 under a recent decline). ESM 6A: A: Ancestral vs. current population size estimated by MSVAR. The red contours give the prior probability density. The cloud of points is the trajectory of the MCMC algorithm which finally settles on a stationary posterior distribution (blue contours). B: Posterior density of time since the population decline. The red dotted line represents the prior, while the blue curve shows the posterior density. ESM 6B: Posterior summaries from MSVAR for population-level parameters (partially pooled across loci) of coalescent simulations of microsatellite evolution in Trigona spinipes. “CurEff” is the current effective population size, “AncEff” the ancestral effective population size, “Mut” the mutation rate, “Time” is the time since population decline, “Mean” is the posterior expectation, “SD” is the posterior std. deviation, “CI x%” is the xth quantile of the posterior, and “Rhat” is the Gelman-Rubin scale-reduction factor. Parameter Mean SD CI 2.5% CI 25% CI 75% CI 97.5% Rhat CurEff (Mean) 3.522 0.586 2.552 3.053 3.982 4.624 1.10 AncEff (Mean) 4.430 0.259 3.926 4.253 4.610 4.936 1.02 Mut (Mean) -3.508 0.243 -4.026 -3.706 -3.366 -3.056 1.00 Time (Mean) 2.831 0.474 1.965 2.496 3.153 3.799 1.06 CurEff (Var) 0.626 0.332 0.047 0.387 0.831 1.342 1.20 AncEff (Var) 0.150 0.117 0.006 0.057 0.217 0.435 1.01 Mut (Var) 0.118 0.095 0.005 0.045 0.168 0.362 1.00 Time (Var) 0.279 0.212 0.012 0.109 0.403 0.782 1.14 ESM 6C: Posterior expectations and 95% credibility intervals for population-level parameters (partially pooled across loci) of current effective population size (CurEff), ancestral effective population size (AncEff), and time since population decline (Time); across priors of varying magnitude. The mutation rate was essentially identical across priors and so is not shown. Prior Mode (log 10) Posterior Estimate (log 10, Mean +/- SD) CurEff AncEff Time CurEff AncEff Time 2 2 2 2.633 +/- 0.465 4.413 +/- 0.264 2.362 +/- 0.643 2 2 4 3.420 +/- 0.479 4.411 +/- 0.267 2.616 +/- 0.475 2 4 2 2.666 +/- 0.535 4.458 +/- 0.258 2.525 +/- 0.599 2 4 4 3.409 +/- 0.435 4.423 +/- 0.261 2.795 +/- 0.611 4 2 2 2.613 +/- 0.553 4.432 +/- 0.263 2.443 +/- 0.520 4 2 4 3.237 +/- 0.510 4.449 +/- 0.257 2.901 +/- 0.426 4 4 2 2.720 +/- 0.439 4.505 +/- 0.261 2.511 +/- 0.502 4 4 4 3.522 +/- 0.586 4.430 +/- 0.259 2.831 +/- 0.474