Supplementary Figure S1. TIC807 coding sequence and flanking
advertisement

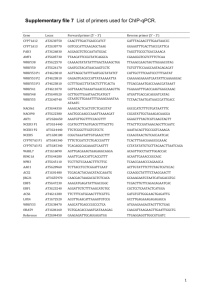
Supplementary Figure S1. TIC807 coding sequence and flanking regions 1 GATCAGAAGCTTTGTAAGCATCTCAGAAGCGTTTTTGCCATCAAAGAAGGGTTAAGGGTA 60 61 TAGCCAGGTCTAAAAGTTTACTAGAAGGGCGAGAGCCGCAATTAAATGAAAGGGAGGAGT 120 121 TCATCAATCAAAAAGAATTGGATATTAGCTAGAGGGTGTTGCAAAACTCATAAATATCCA 180 181 AAAATGAATGCATAAAAAAATCCGTTCTTTTGGAAAAATGATAGTATCCCAAACTATCTC 240 241 CGGAAAGGACGGATTCACTAGTATGATAGCTAACTTTGATCAGAATCAATCCATGTTACA 300 301 GATTTTATTTGCATATGTAGACAGTTTAATCATTGGAGGCGTTCCGCCTTTTACAGGTCG 360 361 TCCACCTATATGTAGAAAGGCTTTATTAAAATGTTTTTTCTTGAAAACTGTATTCCAGAT 420 421 TGATTCTTTACGGAAATTAACTCGTTTTTTACATCAGTATCCTTCATTTCGTGTATCTTG 480 481 TGGCTTTTCTTCACTTCCTCATATCTCTACTTTTTCACGTGTAGGTACTTGGTTTCGTGA 540 541 AGAAGGTATTCCATTGATCCATAAGCAAATTCTTCACGAAATGAATTTCGGTTTAATTCC 600 601 GTGCGTTTTAATTGATCGTACAGCCTTACGGAGTAGTTTATATGATTCTCAAGCAAAATG 660 661 GGGGAAGTCGACTCGTTATGGTTGGTATAAAGGATATAAGGCCCATGTTTGTTCAACACC 720 721 AGAAGGTGTTGTGTTGTCTTACGCGTTTACAACAGCTAACATCCATGATAGCAAAATGGC 780 781 TCCTGTATTGCTTCAAAATATACAAGACCAAAATGTGTTGTTTTCTGTTGCAGATGCTGC 840 841 TTATGATAGTCAGCATATTTATGAAATTGCGCGAGCATGTAATATCTTTGCCATAAATCC 900 901 AATTAATCCAAGGAATGGTGAACAAAATAAGAGTACACATCGTCGTATATTATCTCAATT 960 961 TATACAAACTATCTTAGGTAAGCAGTTGATGAAGGAATGTGGAAAAATTGAACAACAGTT 1020 1021 TAGTAATCTCAAAGATAAAGGGCTGGAACAGCCACGTTGGTATGGAAAAAATCGTTATCT 1080 1081 ATTGCATGTTCAGCTCATTTTTCTGATTCATAACATTGCATATTTATTTTAGTTTTGCAA 1140 1141 CACCCTCAGCTATAAAGAATTAGAAGTGAAGTGAATAGCATGTCATTTCATTTCATTTGT 1200 1201 GTTCATAACTATAAACTATAATTTTATTTTAATGATAAAAGGAGAAATATATTATGCAAA 1260 1261 TTACGCTAGAACAAACTGTAAATGGAAAGAAATTTTTAAATATTAAGAATAAAAATTAAT 1320 1321 ATTTAAAATTGGTTCAAAAAACCAAAAACATACATATATTATTATTGTTGTTTTATTTTT 1380 1381 GAAAATTCAAAAATATATATTTTGTTCATATTTTGTAACGTTAATTATTTTATATTTTAA 1440 1441 ACAATTAGGAGATGATTTTTTTGGCAATTTTAGATTTAAAATCTTTAGTACTCAATGCAA 1500 M A I L D L K S L V L N A I 1501 TAAATTATTGGGGTCCTAAAAATAATAATGGCATACAGGGTGGTGATTTTGGTTACCCTA 1560 N Y W G P K N N N G I Q G G D F G Y P I 1561 TATCAGAAAAACAAATAGATACGTCTATTATAACTTTTACTCATCCTCGTTTAATTCCAT 1620 S E K Q I D T S I I T F T H P R L I P Y 1621 ATGATTTAACAATTCCTCAAAATTTAGAAACTATTTTTACTACAACTCAAGTATTAACAA 1680 D L T I P Q N L E T I F T T T Q V L T N 1681 ATAATACAGATTTACAACAAAGTCAAACTGTTTCTTTTGCTAAAAAAACAACGACAACAA 1740 N T D L Q Q S Q T V S F A K K T T T T T 1741 CTTCAACTTCAACTACAAATGGTTGGACAGAAGGTGGGAAAATTTCAGATACATTAGAAG 1800 S T S T T N G W T E G G K I S D T L E E 1801 AAAAAGTAAGTGTATCTATTCCTTTTATTGGAGAGGGAGGAGGAAAAAACAGTACAACTA 1860 K V S V S I P F I G E G G G K N S T T I 1861 TAGAAGCTAATTTTGCACATAACTCTAGTACTACTACTTTTCAACAGGCTTCAACTGATA 1920 E A N F A H N S S T T T F Q Q A S T D I 1921 TAGAGTGGAATATTTCACAACCAGTATTGGTTCCCCCAAGTAAACAAGTTGTAGCAACAT 1980 E W N I S Q P V L V P P S K Q V V A T L 1981 TAGTTATTATGGGAGGTAATTTTACTATTCCTATGGATTTGATGACTACTATAGATTCTA 2040 V I M G G N F T I P M D L M T T I D S T 2041 CAGAACATTATAGCCATTATAGTGGTTATCCAATATTAACATGGATATCGAGCCCCGATA 2100 E H Y S H Y S G Y P I L T W I S S P D N 2101 ATAGTTATAGTGGTCCATTTATGAGTTGGTATTTTGCAAATTGGCCCAATTTACCATCGG 2160 S Y S G P F M S W Y F A N W P N L P S G 2161 GGTTTGGTCCTTTAAATTCAGATAATACGGTCACTTATACAGGTTCTGTTGTAAGTCAAG 2220 F G P L N S D N T V T Y T G S V V S Q V 2221 TATCAGCTGGTGTATATGCCACTGTACGATTTGATCAATATGATATACACAATTTAAGGA 2280 S A G V Y A T V R F D Q Y D I H N L R T 2281 CAATTGAAAAAACTTGGTATGCACGACATGCAACTCTTCATAATGGAAAGAAAATATCTA 2340 I E K T W Y A R H A T L H N G K K I S I 2341 TAAATAATGTTACTGAAATGGCACCAACAAGTCCAATAAAAACAAATTAAGACTAAAATG 2400 N N V T E M A P T S P I K T N * 2401 AAAAAAGAATAAAAAAATAAAGCTCTTACTTTGTGTATATGATCACAACGTAGGAGCTTT 2460 2461 TTTCATTAGATTTGTTCATACAGCTTTCCTTGTATCCGGGGTAAACCCAGCATCTTCGAG 2520 2521 CAGAATGGACATTTTTAAATTTTCAGACAATTGATAGATTTTTTTATGGTTTTTACGGCA 2580 2581 CGAAATCATAGTTCATGCGTTCTGCGTCGCTTTTTTAAACTGTTAAAACATGAAATGAAC 2640 2641 CTGATGTTTAATTAATTTTGATGAAAAGTGTCACAGAATCATATTTCTATGTCACTTTTT 2700 2701 GTTTAAACTTATTAAATTTACACTGTTAAAATTCTGATTATTATGAAATGTCCGTTTTAC 2760 2761 TTGGAGATGCTGGTTCTACCCCTATCCGCCTGTTTTTATCTTTTAATCGCTTATTGTCAC 2820 2821 TTTCCAATTCTTTGATTATACAGAATAATTTTTCAAATGATTTTCACCTGATATAAGATC 2880 2881 GTACTCCATATGTGATTTATATATATTGTCTATTAGTAAAAATATTTGTGTCCTGTTTTT 2940 2941 ATTTGTTACACGTGATTAAGCATAAAAAGAGCAGAAAAGAGGAAAAAACAATACAACATC 3000 3001 ATAGAGCGAATGATTTATCAATTTTTTCTTGTGTGGGTCCGGTATGCGAAGATGAACTAC 3060 3061 ATCGTTTCCCTTTCTTTATAGCTGCTTCCAATTCAGATTTATCTTCATCTAAAAAAATAG 3120 3121 AGAAGCACTCTTTCCCTATTAATGCAGTACTTTCTTTTCCTAATAACATACTTCCAGCCT 3180 3181 GGTTTATATTTAATACGATACCGTTTGAATCAATCGTTAAAATAGGATC 3229 Supplementary Figure S2. Pair-wise alignment of TIC807 and Cry15Aa1 Program: needle Aligned_sequences: 2 1: Cry15Aa1 2: TIC807 Matrix: EBLOSUM62 Gap_penalty: 10.0 Extend_penalty: 0.5 Length: 376 Identity: Similarity: Gaps: Score: 258.5 Cry15Aa1 TIC807 Cry15Aa1 TIC807 Cry15Aa1 TIC807 Cry15Aa1 TIC807 Cry15Aa1 TIC807 Cry15Aa1 TIC807 Cry15Aa1 96/376 (25.5%) 148/376 (39.4%) 103/376 (27.4%) 1 MAIMN--DIAQDAARAWDIIAGPFIRPGTTPTNRQLFNYQIGNIEVEPGN |||:: .:..:|...| ||....|....: |.|.|...:::... 1 MAILDLKSLVLNAINYW----GPKNNNGIQGGD---FGYPISEKQIDTSI 48 49 LNFS---VVP---------ELDFSVSQDLFNNTSVQQSQTASFNESRTET :.|: ::| |..|:.:|.|.|||.:|||||.||.:..|.| 44 ITFTHPRLIPYDLTIPQNLETIFTTTQVLTNNTDLQQSQTVSFAKKTTTT 86 87 TSTAVTHGVKSGVTVSASAKFNAKILVKSI-----EQTITTTVSTEYNFS |||:.|:|...|..:|.:.:....:.:..| :.:.|...:..:|.| 94 TSTSTTNGWTEGGKISDTLEEKVSVSIPFIGEGGGKNSTTIEANFAHNSS 132 STTTRTNTVTRGWSIAQPVLVPPHSRVTATLQIYKGDFTVPVLL-----S :||.:..:....|:|:|||||||..:|.|||.|..|:||:|:.| | 144 TTTFQQASTDIEWNISQPVLVPPSKQVVATLVIMGGNFTIPMDLMTTIDS 43 93 131 143 176 193 177 LRVYGQTG-----TLAGNPS------FPSLYAATYENTLLGRIREHIAPP ...|.... |...:|. |.|.|.|.:.|. | 194 TEHYSHYSGYPILTWISSPDNSYSGPFMSWYFANWPNL-----------P 215 216 ALFRASNAYISNGVQAIWRGTATTRVSQGLYSVVRIDERPLAGYSGETRT :.|...|: .|.| .:.|:..::||.|:|:.||.|:..:.......:| 233 SGFGPLNS--DNTV--TYTGSVVSQVSAGVYATVRFDQYDIHNLRTIEKT 265 TIC807 266 YYL-PVTLSNSSQILTPGSLGSEIPIINPVPNASCKKENSPIIIHHDREK :|. ..||.| |.:|.|.|....| ..|||..: 279 WYARHATLHN----------GKKISINNVTEMA----PTSPIKTN Cry15Aa1 315 HRERDYDKEHICHDQAEKYERDYDKE 340 TIC807 310 309 232 278 314 309 Supplementary Figure S3. Coding region for a transposase adjacent to the TIC807 gene Query= pMON96219 insert >gb|AAZ99889.1| transposase [Bacillus thuringiensis serovar tenebrionis] Length: 289 E value = e-148 87% sequence identity Query: 263 MIANFDQNQSMLQILFAYVDSLIIGGVPPFTGRPPICRKALLKCFFLKTVFQIDSLRKLT 442 MI NFDQNQS+LQILFAYVDSL IGGVPP TGRPPICRKALLKCFF+KTVFQI+SLRKLT Sbjct: Query: 1 MIINFDQNQSVLQILFAYVDSLTIGGVPPLTGRPPICRKALLKCFFVKTVFQINSLRKLT 60 443 RFLHQYPSFRVSCGFSSLPHISTFSRVGTWFREEGIPLIHKQILHEMNFGLIPCVLIDRT 622 RFLHQYPSFRVSCG S +PHISTFSRVG WFR EGIP+IHKQ L EMN GLIPCVLI+ T Sbjct: 61 RFLHQYPSFRVSCGLSFVPHISTFSRVGNWFRNEGIPMIHKQTLEEMNLGLIPCVLINST 120 Query: 623 ALRSSLYDSQAKWGKSTRYGWYKGYKAHVCSTPEGVVLSYAFTTANIHDSKMAPVLLQNI 802 ALRSSLYDSQAKWGKSTRYGWYKGYKAHVCSTPEG+VLSY+FTT N+H SKMAPVLL++I Sbjct: 121 ALRSSLYDSQAKWGKSTRYGWYKGYKAHVCSTPEGIVLSYSFTTVNVHGSKMAPVLLRDI 180 Query: 803 QDQNVLFSVADAAYDSQHIYEIARACNIFAINPINPRNGEQNKSTHRRILSQFIQTILGK 982 QD+NVLFSVADAAYDSQHIYEIAR NIFA+NPINPRNGEQ KSTHRR+LS F+QT+ GK Sbjct: 181 QDRNVLFSVADAAYDSQHIYEIARISNIFAVNPINPRNGEQIKSTHRRVLSHFVQTLFGK 240 Query: 983 QLMKECGKIEQQFSNLKDKGLEQPRWYGKNRYLLHVQLIFLIHNIAYLF 1129 QLMKE GKIEQQFSNLKDKGLEQPRWYG+NRYLLHVQL+FLIHNIAYLF Sbjct: 241 QLMKERGKIEQQFSNLKDKGLEQPRWYGQNRYLLHVQLVFLIHNIAYLF 289 Supplementary Figure S4. Coding sequence for the plant-expressed TIC807 protein ATGGCTATCCTAGACCTTAAGTCCCTCGTGCTGAACGCCATTAACTACTGGGGCCCTAAGAACA ACAACGGCATCCAGGGCGGTGACTTCGGCTACCCCATCTCTGAGAAGCAGATCGACACTAGCAT CATTACCTTCACCCACCCTCGCTTGATCCCCTACGATCTTACTATCCCGCAGAACCTTGAGACC ATCTTCACCACAACGCAGGTGCTCACCAATAACACTGACCTCCAGCAATCCCAGACCGTGAGCT TTGCGAAGAAGACCACTACCACGACCTCAACTAGCACGACCAACGGTTGGACAGAAGGAGGCAA GATCAGCGACACGCTGGAGGAGAAAGTTTCGGTTAGCATTCCGTTCATCGGTGAGGGTGGCGGG AAGAACTCGACTACCATAGAGGCCAACTTCGCACACAACTCTAGCACCACTACCTTCCAGCAAG CAAGCACTGACATTGAGTGGAACATTAGCCAACCGGTGCTGGTTCCTCCCTCTAAACAAGTTGT CGCGACCCTTGTGATCATGGGAGGCAACTTTACCATCCCTATGGACTTGATGACCACGATTGAT AGTACAGAGCACTACTCCCACTACTCCGGTTACCCTATCCTCACCTGGATCTCGTCCCCAGATA ACTCTTACTCCGGTCCCTTTATGTCATGGTACTTTGCAAACTGGCCTAACCTTCCGAGTGGATT CGGCCCACTGAATAGTGATAACACGGTCACATACACTGGCTCTGTCGTGTCCCAAGTTTCGGCC GGTGTCTACGCTACCGTCCGGTTCGATCAGTATGACATTCACAATCTCCGTACTATCGAGAAGA CTTGGTATGCTCGCCATGCGACGCTGCATAATGGCAAGAAGATTTCTATCAACAATGTCACGGA AATGGCTCCAACATCCCCTATCAAGACAAATTGA TIC807 (5 ng) negative plant ovary anther pistil bract petal leaf boll TIC807 (5 ng) Supplementary Fig. S5. Western blot analysis of floral tissues from a cotton plant transformed with pMON105864. Each lane was loaded with the equivalent of 1 mg fresh tissue. Supplementary Table S1. LC50 determination for Lygus hesperus N Slope + SE LC50 (95% CL) Chi square 262 1.77 + 0.268 72.9 ppm (50.2- 116.4) 43.28 (P<0.001)