ONLINE RESOURCES CAPTIONS Online_Resource_1.pdf
advertisement

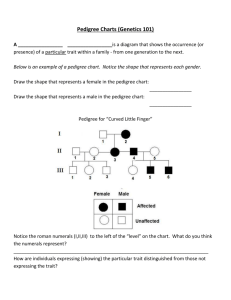
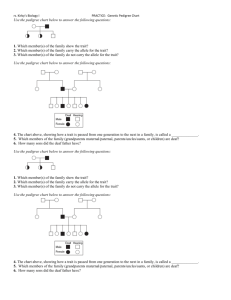
ONLINE RESOURCES CAPTIONS Online_Resource_1.pdf: Supplemental figures and table Fig. S1 The SNP linkage map used for QTL mapping. Fig. S2 Visualization of the pedigree analyzed. A: Horizontal visualization of a representative pedigree of the Processing Peach Breeding Program at UC Davis. B: Horizontal visualization of the progenies with Almond (P. dulcis) ancestry. C: Vertical visualization of progenies with P. argentea ancestry. D: Vertical representation of progenies with P. mira ancestry. E: Vertical visualization of a full-sib family highlighting a male parent introgression line (persXdavidiana = P. persica × P. davidiana). Table S1 Corrected parentages. When dummy# is specified it means that the parent was not available in the analyzed pedigree set. The corrected parentages were corrected based on results from FRANz having a minimum LOD score of 20 and posterior probability > 0.99. In italics are showed the corrected pedigrees for four cultivars. Fig. S3 Determination of the genetic structure for 258 individual members of a pedigree, including peach and almond cultivars and interspecific hybrids, as well as introgression breeding lines. Fig. S4 Genomic breeding values (GBVs) of each trait between years 2011 and 2012 for each accession in the studied pedigree. Fig. S4 Correlation between GBVs of each trait between years 2011 and 2012. Online_Resource_2.pdf: Genomic breeding values per accession per trait per year.