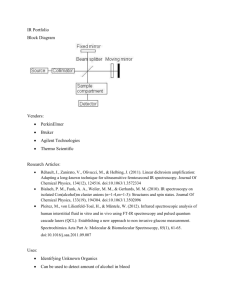
mec13055-sup-0001-AppendixS1
advertisement

APPENDIX S1 Case studies reviewed to describe trends in ecological transcriptomic studies in the last 10 years. The table gives for each of the 575 studies the species, which question categories, whether conducted in the field or lab and whether microarray or RNAseq were included in the study. Question categories: 1 corresponds to studies that investigated the extent of natural variation, 2 corresponds to studies that investigated organismal response to stimulus, and 3 corresponds to studies that investigated the effect of differential gene expression on phenotype. Paper Subject Question Type Field/Lab Technology Abu-Abied et al. 2012 Eucalyptus grandis 1 lab Microarray Aguilar et al. 2010 Anopheles gambiae 1 both Microarray Anderson et al. 2006 Phlebotomus spp. 1 lab Microarray Andersson et al. 2013 multiple 1 field RNAseq Andrés et al. 2013 Gryllus spp. 1 field RNAseq Aranda et al. 2011 Montastraea faveolata 2 lab Microarray Arcà et al. 2005 Anopheles gambiae 1 lab Microarray Arfi et al. 2013 Pycnoporus coccineus 2 lab RNAseq Arnaiz et al. 2010 Paramecium tetraurelia 1 lab Microarray Aubin-Horth et al. 2009 1 field Microarray Avarre et al. 2014 Salmo salar Sarotherodon melanotheron heudelotii 1 lab RNAseq Babik et al. 2010 Myodes glareolus 1 lab RNAseq Bachmann et al. 2012 Lactococcus lactis 2, 3 lab Microarray Bachvaroff et al. 2014 multiple 1 lab RNAseq Badisco et al. 2011 Schistocerca gregaria 1 lab Microarray Badouin et al. 2013 Formica exsecta 1 field RNAseq Bai et al. 2010 Aphis glycines 1 lab RNAseq Bai et al. 2013 Hyriopsis cumingii 1 lab RNAseq Bajgain et al. 2011 Artemisia tridentata 1 lab RNAseq Balakrishnan et al. 2013 Uraeginthus granatina 1 lab RNAseq Baldo et al. 2011 multiple 1 lab RNAseq Ballerini et al. 2013 Iris fulva 1 lab RNAseq Banni et al. 2011 Mytilus galloprovincialis 1 lab Microarray 1 Bao et al. 2012 Nilaparvata lugens 1 lab RNAseq Bao et al. 2013 Nilaparvata lugens 2 lab RNAseq Bar-Yaacov et al. 2013 Chamaeleo chamaeleon 1 field RNAseq Barah et al. 2013 Arabidopsis thaliana 2 lab Microarray Bardil et al. 2011 Coffee arabica 1, 2 lab Microarray Barreto et al. 2011 Tigriopus californicus 1 lab RNAseq Barshis et al. 2013 Acropora hyacinthus 1, 2 lab RNAseq Bass et al. 2012 Nilaparvata lugens 1 lab RNAseq Bassene et al. 2010 1 field Microarray Baumgarten et al. 2013 Citrus spp. Symbiodinium microadriaticum 2 lab RNAseq Bay et al. 2009 Acropora millepora 1 lab Microarray Bay et al. 2009b Acropora millepora 1 lab Microarray Bayne et al. 2006 Oncorhynchus mykiss 2 lab Microarray Bazinet et al. 2013 multiple 1 lab RNAseq Becks et al. 2012 multiple 1, 2 lab Microarray Beemster et al. 2005 Arabidopsis thaliana 1 lab Microarray Belgard et al. 2013 multiple 1 lab RNAseq Bermingham et al. 2009 Buchnera aphidicola 1 lab Microarray Bernatchez et al. 2011 Anguilla spp. 1 field Microarray Bettencourt et al. 2010 Bathymodiuolus azoricus 1 field RNAseq Bilyk and Cheng 2013 Pagothenia borchgrevinki 1 lab RNAseq Boltana et al. 2013 Danio rerio 2, 3 lab Microarray Bonneau et al. 2013 Medicago truncatula 2, 3 lab Microarray Bonneaud et al. 2011 Carpodacus mexicanus 1, 2 lab Microarray Bougas et al. 2010 Salvelinus fontinalis 1 lab Microarray Bougas et al. 2013 Salvelinus fontinalis 1 lab Microarray Boulet et al. 2012 Salvelinus fontinalis 1, 3 lab Microarray Brady et al. 2011 Acropora millepora 1 lab RNAseq Brawand et al. 2011 multiple 1, 3 lab RNAseq Briscoe et al. 2013 Broekgaarden et al. 2008 Broekgaarden et al. 2011 Heliconius melpomene 1 lab RNAseq Brassica oleracea 2 lab Microarray Brassica nigra 2 lab Microarray Bruce et al. 2008 Arabidopsis thaliana 2, 3 lab Microarray Bryk et al. 2013 Mus musculus 1 lab Microarray Buckley et al. 2006 Gillichthys mirabilis 2 lab Microarray 2 Burke and Moran 2011 Acyrthosiphon pisum 1, 2 lab RNAseq Burke and Strand 2014 Microplitis demolitor 3 lab RNAseq Burki et al. 2013 Mikrocytos mackini 1 lab RNAseq Burns et al. 2012 Amphiura filiformis 2, 3 field Microarray Burns et al. 2013 Ophionotus victoriae 1, 2 lab RNAseq Busby et al. 2011 Saccharomyces spp. 1 lab RNAseq Calla et al. 2014 Ceratitis capitata 2 both RNAseq Campanero et al. 2005 Photobacterium profundum 1 lab Microarray Cardoen et al. 2011 Apis mellifera carnica 3 lab Microarray Carvalhais et al. 2013 Bacillus amyloliquefaciens 2 lab Microarray Cassone et al. 2011 Anopheles gambiae 2 lab Microarray Castilho et al. 2009 Thunnus orientalis 2 lab Microarray Castoe et al. 2013 Celorio-Mancera et al. 2013 Python molurus bivittatus 1 lab RNAseq Polygonia c-album 2 lab RNAseq Chan et al. 2012 Chaney and Gracey 2011 Arabidopsis thaliana 2, 3 lab Microarray Crassostrea gigas 2 lab Microarray Chapman et al. 2009 Crassostrea virginica 2 field Microarray Chapman et al. 2011 Crassostrea virginica 2 field Microarray Chapman et al. 2013 Senecio spp. 1 lab RNAseq Chaudhary et al. 2008 Gossypium barbadense 2 lab Microarray Chelaifa et al. 2010 Spartina spp. 1 lab Microarray Chelaifa et al. 2010b Spartina spp. 1 lab Microarray Chen et al. 2010 Locusta migratoria 1 lab RNAseq Chen et al. 2011 Balanus amphitrite 1 lab RNAseq Chen et al. 2013 Vitis vinifera 1 lab Microarray Chen, M. et al. 2012 1 lab RNAseq Chen, Z. et al. 2012 multiple Methanolobus psychrophilus R15 2 lab RNAseq Cheviron et al. 2008 Zonotrichia capensis 1, 2 field Microarray Cheviron et al. 2014 Peromyscus maniculatus 1, 3 both RNAseq Chiari et al. 2012 1 lab RNAseq Chu et al. 2012 multiple Paradoxornis webbianus bulomachus 1 field RNAseq Chun et al. 2008 Euprymna scolopes 2 field Microarray Clark et al. 2010 Laternula elliptica 2 lab Microarray Clark et al. 2012 Desulfovibrio vulgaris 1, 3 lab Microarray Coate and Doyle 2010 Glycine dolichocarpa 1 lab Microarray 3 Coate et al. 2013 Glycine dolichocarpa 1, 3 lab RNAseq Cohen et al. 2007 multiple 1 lab Microarray Collén et al. 2007 Chondrus crispus 2 lab Microarray Colombo et al. 2013 multiple 1, 3 field RNAseq Comeault et al. 2012 Timema cristinae 1 field RNAseq Conaco et al. 2012 Conant and Wagner 2004 Connor and Gracey 2011 Amphimedon queenslandica 1 field RNAseq Caenorhabditis elegans 3 lab Microarray Mytilus californianus 1 both Microarray Cook et al. 2011 Dolerus spp. 1 field RNAseq Coppe et al. 2010 Corredor-Adámez et al. 2005 Anguilla anguilla 1 field RNAseq Danio rerio 1 lab Microarray Cseke et al. 2009 Populus tremuloides 1 field Microarray Cui et al. 2013 multiple 1 lab RNAseq Cullingham et al. 2013 Pinus spp. 1 field RNAseq Czesny et al. 2012 Alosa pseudoharengus 1 field RNAseq Czypionka et al. 2012 Cottus spp. 1 lab Microarray Davidson et al. 2012 multiple 1 lab RNAseq de Boer et al. 2011 Folsomia candida 2 field Microarray de Carvalho et al. 2013 De Wit and Palumbi 2013 Spartina spp. 1 lab RNAseq Haliotis rufescens 1 field RNAseq Debes et al. 2012 Salmo salar 1, 2 lab Microarray Denekamp et al. 2011 Brachionus plicatilis 1 lab RNAseq Denver et al. 2005 Caenorhabditis elegans 1 lab Microarray Der et al. 2011 Derome and Bernatchez 2006 Pteridium aquilinum 1 lab RNAseq Coregonus artedi 1 field Microarray Derome et al. 2006 Coregonus clupeaformis 1 field Microarray Derome et al. 2008 Coregonus spp. 1, 3 lab Microarray Des Marais et al. 2012 Arabidopsis thaliana 1, 2 lab Microarray DeSalvo et al. 2008 Montastraea faveolata 2 lab Microarray DeSalvo et al. 2010 Acropora palmata 2 field Microarray DeSalvo et al. 2010b Montastraea faveolata 1, 2 lab Microarray Dhar et al. 2011 Domazet-Loso and Tautz, et al. 2010 Saccharomyces cerevisiae 2, 3 lab Microarray Danio rerio 1 lab Microarray Doroszuk et al. 2012 Drosophila melanogaster 1, 3 lab Microarray 4 dos Santos Ferreira et al. 2007 Euglena gracilis 2 lab Microarray Dou et al. 2013 Liposcelis bostrychophila 1 lab RNAseq Douglas et al. 2011 Buchnera aphidicola 2 lab Microarray Dowen et al. 2012 Arabidopsis thaliana 2, 3 lab RNAseq Drew et al. 2012 Danio rerio 1, 3 lab Microarray Dudycha et al. 2012 Daphnia spp. 1 lab Microarray Dunning et al. 2013 Micrarchus nov. sp. 2 2 lab RNAseq Durban et al. 2011 multiple 1 lab RNAseq Dworkin et al. 2009 Drosophila melanogaster 1 lab Microarray Ekblom et al. 2012 Philomachus pugnax 1, 3 lab RNAseq Elmer et al. 2010 Amphilophus spp. 1 lab RNAseq Etebari et al. 2011 Plutella xylostella 2 lab RNAseq Evans et al. 2008 Gillichthys mirabilis 2 lab Microarray Evans et al. 2011 1, 2 field Microarray Evans et al. 2013 Ewen-Campen et al. 2011 Oncorhynchus nerka Strongylocentrotus purpuratus 2 lab Microarray Oncopeltus fasciatus 1 lab RNAseq Fabro et al. 2008 Arabidopsis thaliana 2 lab Microarray Fan et al. 2012 Bacillus amyloliquefaciens 2 lab Microarray Fan et al. 2013 Salicornia europaea 2 lab RNAseq Fang et al. 2013 Caulobacter crescentus 1 lab RNAseq Fehsenfeld et al. 2011 Carcinus maenas 2 lab Microarray Feldmeyer et al. 2014 Temnothorax longispinosus 1, 3 lab RNAseq Ferreira et al. 2013 Polistes canadensis 1, 3 field RNAseq Filteau et al. 2013 Coregonus cupleaformis 1 lab Microarray Fonsesca et al. 2011 Pseudomonas putida 2 lab Microarray Frangipani et al. 2014 Pseudomonas aeruginosa 3 lab Microarray Freitak et al. 2012 Tribolium castaneum 1, 2 lab Microarray Friedrich et al. 2011 Ptomaphagus hirtus 1 lab RNAseq Fu and He 2012 Hypophthalmichthus molitrix 1 field RNAseq Gallagher et al. 2008 Oncorhynchus kisutch 1 lab Microarray Galletti et al. 2013 Rickettsia rickettsii 2 lab Microarray Ganot et al. 2011 Anemonia viridis 1 lab Microarray Garcia et al. 2012 Garcia-Vedrenne et al. 2013 Drosophila melanogaster 2, 3 lab Microarray Quahog Parasite Unknown 2 lab RNAseq Genissel et al. 2008 Drosophila melanogaster 1 lab Microarray 5 Geraldes et al. 2011 Populus trichocarpa 1 lab RNAseq Gierga et al. 2012 Synechoccus spp. 1, 2 lab Microarray Giger et al. 2008 Salmo spp. 1 field Microarray Gioti et al. 2012 Drosophila melanogaster 1, 3 lab Microarray Gioti et al. 2013 Neurospora spp. 1 lab RNAseq Goetz et al. 2010 Salvelinus namaycush 1, 3 lab RNAseq Goltsev et al. 2009 Anopheles gambiae 1 lab Microarray Gomulski et al. 2012 Ceratitis capitata 1 lab Microarray Gracey et al. 2004 Cyprinus carpio 2 lab Microarray Gracey et al. 2008 Granados-Cifuentes et al. 2013 Mytilus californianus 2 field Microarray Acropora millepora 1 lab Microarray Gross, J. et al. 2013 Astyanax mexicanus 1 lab RNAseq Gross, S. et al. 2013 Grostern and AlvarezCohen 2013 Agave spp. Pseudonocardia dioxanivorans 1 field RNAseq 2 lab Microarray Grumaz et al. 2013 Candida spp. 1 lab RNAseq Gu et al. 2013 Aphis gossypii 1, 2, 3 lab RNAseq Guggisberg et al. 2013 Cirsium arvense L. 1, 2 lab Microarray Gui et al. 2013 Sousa chinensis 1 field RNAseq Gunter et al. 2013 Astatoreochromis alluaudi 1, 2 lab RNAseq Guo et al. 2013 Pungitius pungitius 1 lab RNAseq Hall et al. 2013 Pinus spp. 1 field RNAseq Hanson et al. 2013 Brachionus calyciflorus 1 lab RNAseq Harms et al. 2013 Hyas araneus 1 lab RNAseq Harr et al. 2010 Mus spp. 1 lab RNAseq Harris et al. 2013 Peromyscus leucopus 1 field RNAseq He et al. 2010 Desulfovibrio vulgaris 2, 3 lab Microarray Healy et al. 2010 Fundulus heteroclitus 2 field Microarray Heath et al. 2012 Medicago truncatula 1 lab Microarray Hegarty et al. 2005 Senecio spp. 1 lab Microarray Hegarty et al. 2009 Senecio spp. 1, 2 lab Microarray Henning et al. 2013 Amphilophus citrinellus 1, 3 lab RNAseq Herranz et al. 2013 Drosopihila melanogaster 2 lab Microarray Hershkovitz et al. 2013 Metschnikowia fructicola 1, 2 lab RNAseq Hewson et al. 2009 Crocosphaera watsonii 1 field RNAseq Hines et al. 2012 Heliconius erato 1, 3 lab Microarray Hocher et al. 2011 multiple 1 lab Microarray 6 Hodgins et al. 2014 Ambrosia artemisiifolia 1, 2 field Microarray Hoffman et al. 2011 Arctocephalus gazella 1 field RNAseq Hohnjec et al. 2005 Medicago truncatula 1, 2 lab Microarray Holliday et al. 2008 Picea sitchensis 1, 2 lab Microarray Hori et al. 2013 Gadus morhua 2 lab Microarray Horvarth et al. 2005 Euphorbia esula 2 lab Microarray Hou et al. 2012 Loxodonta africana 1 lab Microarray Hoy et al. 2013 Metaseiulus occidentalis 1 lab RNAseq Hull et al. 2013 Lygus hesperus 1 lab RNAseq Hüning et al. 2013 Mytilus edulis 2 lab RNAseq Huth and Place 2013 Trematomus bernacchii 2 lab RNAseq Hwang et al. 2008 Ibarra-Laclette et al. 2011 Oncorhynchus kisutch 2 lab Microarray Utricularia gibba 1 field RNAseq Ignatov et al. 2010 Immonen and Ritchie 2012 Mycobacterium avium 1 lab RNAseq 2 lab Microarray Isanapong et al. 2013 Drosophila melanogaster Diplosphaera colotermitum TAV2 1 lab Microarray Jammes et al. 2005 Arabidopsis thaliana 2 lab Microarray Jansen et al. 2013 Daphnia magna 2 lab Microarray Janz et al. 2010 Populus spp. 2, 3 lab Microarray Jeffries et al. 2012 Oncorhynchus nerka 2 lab Microarray Jeffries et al. 2014 Oncorhyncus spp. 1, 2 lab RNAseq Jeukens et al. 2010 Coregonus clupeaformis 1 field RNAseq Jeyasingh et al. 2011 Daphnia pulex 2, 3 lab Microarray Jiang et al. 2009 Jiménez-Guri et al. 2013 Johansson, F. et al. 2013 Johansson, H. et al. 2013 Drosophila spp. 1 lab Microarray multiple 1 lab RNAseq Rana temporaria 2, 3 lab Microarray Formica exsecta 1 lab RNAseq Johnson et al. 2012 Karenia brevis 1, 2 lab Microarray Jouaux et al. 2013 Crassostrea gigas 1, 2 field Microarray Kaakoush et al. 2009 multiple 2 lab Microarray Kadam et al. 2008 Myxococcus xanthus 1, 3 lab Microarray Kalyuzhnaya et al. 2010 Methylotenera mobilis 1 lab Microarray Kanter et al. 2013 Ambrosia artemisiifolia 2, 3 lab RNAseq Kao et al. 2008 Saccharomyces cerevisiae 1, 3 lab Microarray 7 Kao et al. 2013 Schmidtea mediterranea 1 lab RNAseq Kaplinsky et al. 2013 Trachemys scripta 1 lab RNAseq Karkiuki et al. 2010 Brugia spp 1 lab Microarray Kassahn et al. 2007 Pomecentrus moluccensis 2 lab Microarray Katoh et al. 2004 Anabaena 2 lab Microarray Kawanabe et al. 2012 Arabidopsis spp. 1 lab Microarray Keeling et al. 2012 Dendoctonus ponderosae 1 field RNAseq Kegel et al. 2013 Emiliania huxleyi 1, 3 lab Microarray Kelley et al. 2012 Poecilia mexicana 1 field RNAseq Kempema et al. 2007 Arabidopsis thaliana 2 lab Microarray Kersten et al. 2013 2 lab RNAseq Khaitovich et al. 2005 Quercus rober L. Pan troglodytes, Homo sapiens 1 lab Microarray Kijimoto et al. 2009 Onthophagus taurus 1 lab Microarray Kim et al. 2010 Dunaliella salina 2 lab Microarray Kim et al. 2013 Pseudomonas putida 2 lab RNAseq Kimura et al. 2011 Dokdonia sp. MED134 2 lab RNAseq Kitano et al. 2011 Gasterosteus aculeatus 1, 3 lab Microarray Kobayashi et al. 2013 Shorea beccariana 2, 3 field RNAseq Konotchick et al. 2013 Macrocystis pyrifera 1 field RNAseq Kozak et al. 2014 Lucania spp. 1, 3 lab RNAseq Kunster et al. 2010 multiple 1 lab RNAseq Kusnierczyk et al. 2008 2, 3 lab Microarray Küster et al. 2007 Arabidopsis thaliana Medicago truncatula, Glomus intraradices, Amanita muscaria 1 lab Microarray Kvist et al. 2013 Meliteaea cinxia 1, 2 lab Microarray Lai et al. 2008 Helianthus annus 1, 2 lab Microarray Lang et al. 2009 Crassostrea gigas 1, 2 lab Microarray Latta et al. 2012 Daphnia pulex 2 lab Microarray Le Trionnaire et al. 2009 Acyrthosiphon pisum 1, 3 lab Microarray Le Trionnaire et al. 2012 Acyrthosiphon pisum 1 lab Microarray Leakey et al. 2009 Glycine max 1, 3 field Microarray Lee et al. 2011 Dehalococcoides spp. 3 lab Microarray Lee et al. 2013 Laodelphax striatellus 1 lab RNAseq Lehnert et al. 2012 Aiptasia pallida 1 lab RNAseq Lei et al. 2014 Plutella xylostella 2 lab RNAseq Lemay et al. 2013 Oncorhynchus nerka 1 field RNAseq 8 Lemay et al. 2013b Lemmetyinen et al. 2013 Lewandowska-Sabat et al. 2012 Ochotona princeps 1 field RNAseq Salmo salar 1 lab Microarray Arabidopsis thaliana 1 field Microarray Li and Brouwer 2013 Palaemonetes pugio 2, 3 field Microarray Li et al. 2013 Chrysopa pallens 1 lab RNAseq Li, A. et al. 2012 Larix spp. 1 field Microarray Li, BW et al. 2012 Brugia malayi 1 lab Microarray Li, J. et al. 2012 Helicoverpa armigera 1 lab RNAseq Li, S. et al. 2012 Cnaphalocrocis medinalis 1 lab RNAseq Lin et al. 2014 Tetraclita japonica formosana 1, 3 field RNAseq Liu et al. 2012 Aphis glycines 1 lab RNAseq Liu, G. et al. 2013 multiple 1 lab RNAseq Liu, Z. et al. 2013 Lockwood and Somero 2011 Vicia sativa 1 lab RNAseq Mytilus spp. 1, 2 lab Microarray Lockwood et al. 2010 Mytilus spp. 1, 2 lab Microarray Logacheva et al. 2011 Fagopyrum spp. 1 lab RNAseq Lorenz et al. 2010 Pinus taeda 2 lab Microarray Lowe et al. 2011 2 lab RNAseq Ludeman et al. 2014 Oxyrrhis marina Ephydatia muelleri, Spongilla lacustris 3 lab RNAseq Lundberg et al. 2013 Phylloscopus trochilus 1 field RNAseq Lv et al. 2013 Portunus trituberculatus 2 lab RNAseq Maaty et al. 2009 Sulfolobus solfataricus 2, 3 lab Microarray Malenke et al. 2013 Neotoma lepida 1, 2 lab Microarray Malone et al. 2006 Xenopus spp. 1 both Microarray Mancia et al. 2008 Tursiops truncatus 2 field Microarray Mancia et al. 2010 Tursiops truncatus 1 field Microarray Mancia et al. 2012 Zalophus californianus 2, 3 field Microarray Mane et al. 2007 Arabidopsis thaliana 2 lab Microarray Manfredini et al. 2014 Solenopsis invicta 1, 2 lab Microarray Manousaki et al. 2013 Amphilophus citrinellus 1, 3 field RNAseq Margres et al. 2013 Micrurus fulvius 1 lab RNAseq Marinković et al. 2012 Chironomus riparius 1 lab RNAseq Marques et al. 2008 Martens-Uzunova et al. 2008 Danio rerio 2, 3 lab Microarray Aspergillus niger 1, 3 lab Microarray 9 Martin et al. 2006 Salmo salar 2 lab Microarray Martinez et al. 2009 Postia placenta 3 lab Microarray Mattila et al. 2012 Stegodyphus spp. 1 lab RNAseq Matz et al. 2008 Pseudomonas aeruginosa 2 field Microarray Matzkin 2012 Drosophila mojavensis 1 lab Microarray Maughan et al. 2009 Bacillus subtilis 3 lab Microarray Mauter et al. 2013 Escherichia coli 2 lab Microarray Mavarez et al. 2009 Salvelinus fontinalis 1 lab Microarray McCoy et al. 2014 Euphydryas gillettii 1 field RNAseq McDonnell et al. 2013 Apis mellifera 2 lab RNAseq McKain et al. 2012 multiple 1 lab RNAseq Meier et al. 2014 Salmo trutta 3 field Microarray Mela et al. 2011 multiple 2, 3 lab Microarray Meng et al. 2013 Sinocyclocheilus spp. 1 lab RNAseq Meyer et al. 2011 Acropora millepora 2 lab RNAseq Mezey et al. 2008 Drosophila spp. 1 lab Microarray Michely et al. 2013 Ostreococcus tauri 1 lab Microarray Milan et al. 2011 Ruditapes phillippinarum 1, 2 lab RNAseq Milan et al. 2013 Ruditapes phillippinarum 1, 2 field Microarray Miller et al. 2012 Sphenodon punctatus 1 lab RNAseq Mittapalli et al. 2010 Agrilus planipennis 1 field RNAseq Mok et al. 2007 Plasmodium falciparum 1 lab Microarray Moreno-Paz et al. 2010 Leptospirillum spp. 1 field Microarray Moriwaki et al. 2010 Arabidopsis thaliana 2 lab Microarray Moya et al. 2012 Moyers and Rieseberg 2013 Mukhopadhyay et al. 2006 Anemonia viridis 2 lab Microarray Helianthus annuus 1, 3 field RNAseq Desulfovibrio vulgaris 2 lab Microarray Nagano et al. 2012 Oryza sativa 1, 2 field Microarray Natt et al. 2012 Gallus gallus 1 lab Microarray Navarro et al. 2013 Dreissena polymorpha 1, 2, 3 field Microarray Newton et al. 2013 Lates calcarifer 1, 2, 3 lab RNAseq Nguyen et al. 2012 Helicoverpa zea 1 lab RNAseq Nikinmaa et al. 2013 Nipitwattanaphon et al. 2013 Gasterosteus aculeatus 3 lab Microarray Solenopsis invicta 1 lab Microarray Niu, D. et al. 2013 Sinonovacula constricta 1 field RNAseq 10 Niu, S. et al. 2013 Pinus tabuliformis 1 lab RNAseq Norman et al. 2014 Salvelinus alpinus 1, 2, 3 lab RNAseq Nota et al. 2010 Folsomia candida 2 lab Microarray Novo et al. 2013 multiple 1 field RNAseq Nowack et al. 2011 Paulinella chomatophora 1 lab RNAseq Nyberg et al.2012 Pristina leidyi 1 lab RNAseq Nygren et al. 2012 1 lab Microarray O'Donnell et al. 2010 Neurospora intermedia Strongylocentrotus purpuratus 2 lab Microarray Ometto et al. 2012 Solenopsis spp. 1 lab Microarray Ometto et al. 2013 Drosophila suzukii 1 lab RNAseq Osada et al. 2006 Drosophila melanogaster 1 lab Microarray Overgaard et al. 2010 Drosophila melanogaster 2 lab Microarray Ozawa et al. 2012 Padilla-Gamiño et al. 2013 multiple Strongylocentrotus purpuratus. 2, 3 lab RNAseq 2, 3 lab Microarray Pagarete et al. 2011 multiple 2 field Microarray Page et al. 2013 Ambystoma spp. 1 lab Microarray Pallavicini et al. 2013 Latimeria menadoensis 1 lab RNAseq Palumbi et al. 2014 Acropora hyacinthus 1, 2 both RNAseq Pandey et al. 2008 Nicotiana attenuata 2, 3 lab RNAseq Parikh et al. 2010 Dictyostelium spp. 1 lab RNAseq Park et al. 2008 Populus deltoides 1 lab Microarray Patell et al. 2010 Pseudomonas aeruginosa 1 lab Microarray Patzelt et al. 2013 Dinoroseobacter shibae 3 lab Microarray Pauchet et al. 2009 Chrysomela tremulae 1 lab RNAseq Pavey et al. 2011 Oncorhynchus nerka 1 field Microarray Pellino et al. 2013 Ranunculus auricomus spp. 1 field RNAseq Pellizzari et al. 2013 Sparus aurata 2 lab Microarray Peng et al. 2011 Nilaparvata lugens 1 lab RNAseq Perdiguero et al. 2013 Pinus pinaster 2 lab Microarray Perez-Porro et al. 2013 Crella elegans 1 field RNAseq Philipp et al. 2012 2 lab RNAseq Philippe et al. 2010 Mytilus edulis Populus trichocarpa x deltoides 2, 3 lab Microarray Pierce et al. 2012 Elysia chlorotica 1 lab RNAseq Pina et al. 2005 Arabidopsis thaliana 1 lab Microarray Piveteau et al. 2011 Listeria monocytogenes 1, 2 lab Microarray 11 Place et al. 2012 Mytilus californianus 1, 2 field Microarray Plett et al. 2010 Populus trumula x alba 1 lab Microarray Poelchau et al. 2011 Aedes albopictus 1 lab RNAseq Poelchau et al. 2013 Aedes albopictus 1, 2 lab RNAseq Polato et al. 2010 Montastraea faveolata 2 lab Microarray Polato et al. 2011 Acropora palmata 1, 2 lab RNAseq Polato et al. 2013 Acropora palmata 1, 2 field Microarray Pontes et al. 2008 Sodalis glossinidius 1 lab Microarray Porcelli et al. 2013 Campylobacter jejuni 1 lab RNAseq Portune et al. 2010 Acropora palmata 1, 2 lab Microarray Pravosudov et al. 2013 Poecile atricapillus 1, 3 lab RNAseq Prince et al. 2010 Tribolium castaneum 1 lab Microarray Prosdocimi et al. 2011 multiple 1 field RNAseq Provost et al. 2011 Spodoptera frugiperda 2, 3 lab Microarray Qiao et al. 2013 Odorrana margaretae 1 field RNAseq Rajpurohit et al. 2013 Renaut and Bernatchez 2011 Drosophila mojavensis 1, 2 lab Microarray Coregonus spp. 1, 3 lab Microarray Renaut et al. 2009 Coregonus cupleaformis 1 lab Microarray Renaut et al. 2010 Coregonus spp. 1 lab RNAseq Renaut et al. 2011 Coregonus clupeaformis 1 field RNAseq Renaut et al. 2014 Rendón-Anaya et al. 2012 Helianthus spp. Centruroides noxius Hoffmann 1 lab RNAseq 1 lab RNAseq Rensink et al. 2005 Solanacae family 1 lab Microarray Rey et al. 2013 Danio rerio 1, 3 lab Microarray Ribeiro et al. 2012 Antricola delacruzi 1 field RNAseq Richards et al. 2012 Arabidopsis thaliana 1, 2 field Microarray Richier et al. 2008 Anthopleura elegantissima 2 lab Microarray Riley et al. 2014 Rhizophagus spp. 1 lab RNAseq Rinker et al. 2013 Anopheles spp. 3 lab RNAseq Roberge et al. 2007 Salmo salar 1 lab Microarray Roberge et al. 2008 Salmo salar Lithobates clamitans, Pseudacris regilla 2 lab Microarray 1 lab RNAseq Populus trichocarpa 1 field Microarray Anthopleura elegantissima 2 lab Microarray Robertson et al. 2014 Rodgers-Melnick et al. 2012 Rodriguez-Lanetty et al. 2006 12 Roeding et al. 2009 Pandinus imperator 1 lab RNAseq Rokitta et al. 2011 Emiliana huxleyi 1 lab Microarray Ronges et al. 2012 Petrolisthes cinctipes 2, 3 lab Microarray Rosenblum et al. 2012 Rana spp. 2 lab Microarray Salcedo et al. 2012 Heterocapsa circularisquama 1 lab RNAseq Salem et al. 2006 Oncorhynchus mykiss 1 lab Microarray Samils et al. 2013 Neurospora tetrasperma 1 lab Microarray Sanogo et al. 2011 Gasterosteus aculeatus 2 lab Microarray Sanogo et al. 2012 Gasterosteus aculeatus 1, 2 lab Microarray Santos et al. 2008 Danio rerio 1 lab Microarray Sass et al. 2013 Burkholderia cenocepacia 2, 3 lab Microarray Sauvage et al. 2010 Salvelinus fontinalis 2 lab Microarray Schmidt et al. 2013 Schnitzler and Weis 2010 Chironomus spp. 1 lab RNAseq Fungia scutaria 1, 2 lab Microarray Schoville et al. 2012 Tigriopus californicus 1 lab RNAseq Schramm et al. 2006 Arabidopsis thaliana 2 lab Microarray Schwartz, J. et al. 2010 Elysia chlorotica 1 lab RNAseq Schwartz, T. et al. 2010 Thamnophis elegans 1 both RNAseq Schweizer et al. 2013 Arabidopsis thaliana 2 lab Microarray Scully et al. 2013 Anoplophora glabripennis 1 lab RNAseq Seeb et al. 2011 Sellin Jeffries et al. 2012 Oncorhynchus keta 1 field RNAseq Pimephales promelas 2 field Microarray Sen et al. 2013 Reticulitermes flavipes 2 lab Microarray Seo et al. 2012 multiple 2 lab Microarray Seto and Tamura 2013 Drosophila spp. 1, 2 lab RNAseq Shaw et al. 2012 Artibeus jamaicensis 1 lab RNAseq Shi et al. 2013 Reaumuria soongorica 1 field RNAseq Shiao et al. 2012 Mus musculus 1 lab RNAseq Shin et al. 2012 multiple 1 lab RNAseq Sikhakolli et al. 2012 Fusarium spp. 1 lab RNAseq Sinha et al. 2012 multiple 1 lab Microarray Sloan et al. 2012 Silene vulgaris 1 lab RNAseq Small et al. 2009 Danio rerio 1 lab Microarray Small et al. 2013 Syngnathus spp. 1 lab RNAseq Smith, G. et al. 2013 Drosophila mojavensis 2, 3 lab RNAseq Smith, S. et al. 2013 Melanotaenia duboulayi 1, 2 lab RNAseq 13 Sneddon et al. 2005 Oncorhynchus mykiss 1 lab Microarray Snell-Rood et al. 2010 Onthophagus taurus 1 lab Microarray Soós et al. 2012 Lactuca sativa 2, 3 lab Microarray Spiess et al. 2012 Quercus robur 2, 3 lab Microarray Srinivasan et al. 2013 Panagrellus redivivus 1 lab RNAseq StCyr et al. 2008 Coregonus spp. 1, 3 both Microarray Stillman et al. 2009 Petrolisthes cinctipes 2 lab Microarray Stoll et al. 2009 Blochmannia floridanus 1 lab Microarray Su et al. 2013 Phalaenopsis aphrodite 3 lab Microarray Sun et al. 2012 Pomacea canaliculata 1 lab RNAseq Sussarellu et al. 2010 Crassostrea gigas 2 lab Microarray Sutherland et al. 2012 Lepeophtheirus salmonis 2 lab Microarray Szoevenyi et al. 2011 Funaria hygrometrica 1 lab RNAseq Szoevenyi et al. 2013 Funaria hygrometrica 1 lab RNAseq Tatematsu et al. 2005 Arabidopsis thaliana 1 lab Microarray Taylor et al. 2005 Populus x euramericana 2 lab Microarray Teo et al. 2009 Gracilaria changii 2 lab Microarray Teulier et al. 2012 Aptenodytes patagonicus 1, 3 field Microarray Thompson et al. 2011 Prochlorococcus spp. 2 lab Microarray Timmerhaus et al. 2012 Salmo salar 2 lab Microarray Timmermans et al. 2009 Timmins-Schiffman et al. 2013 Folsomia candida 2 lab Microarray multiple 1 lab RNAseq Tisch and Schmoll 2013 Trichoderma reesei 2 lab Microarray Tisserant et al. 2011 Tuber melanosporum 1 lab RNAseq Tisserant et al. 2012 Todgeham and Hofmann 2009 Toffano-Nioche et al. 2012 Glomus intraradices Strongylocentrotus purpuratus 1 lab RNAseq 2 lab Microarray Vibrio splendidus 1 lab RNAseq Tomasch et al. 2011 Dinoroseobacter shibae 2 lab Microarray Toth et al. 2010 Polistes metricus 1 field Microarray Toullec et al. 2013 Euphausia crystallorophias 1 field RNAseq Traeger et al. 2013 multiple 1, 2 lab RNAseq Travers et al. 2010 Andropogon gerardii 2 field Microarray 14 Traylor-Knowles et al. 2011 Pocillopora damicornis 1, 2 lab RNAseq Truebano et al. 2010 Tsagkogeorga et al. 2010 Latemula elliptica 2 lab Microarray Microcosmus squamiger 1 field RNAseq Tymchuk et al. 2010 Salmo salar 1 both Microarray Tzika et al. 2011 multiple 1 lab RNAseq Uebbing et al. 2013 Ficedula albicollis 1 field RNAseq Unal et al. 2013 Calanus finmarchicus 1 field Microarray Valles et al. 2012 van de Mortel et al. 2012 Nylanderia pubens 1 lab RNAseq Arabidopsis thaliana 2, 3 lab RNAseq Thoracosphaera heimii Folsomia candida, Protaphorura fimata 2 lab RNAseq 2 lab Microarray Azospirillum brasilense 2 lab Microarray Gryllus firmus 1, 3 lab RNAseq Vences et al. 2013 Rana spp. 1 field RNAseq Venier et al. 2006 Mytilus galloprovincialis 2 lab Microarray Vera et al. 2008 Melitaea cinxia 1 field RNAseq Vercruysse et al. 2011 Rhizobium etli 2, 3 lab Microarray Vermeulen et al. 2013 Drosophila melanogaster 1, 2 lab Microarray Verta et al. 2013 Picea glauca 1 field Microarray Vesala et al. 2012 Drosophila spp. 2 lab Microarray Vezzi et al. 2005 Photobacterium profundum 1 lab Microarray Vicoso et al. 2013 multiple 1 both RNAseq Vidal-Dupiol et al. 2013 Voelckel and Baldwin 2004 Pocillopora damicornis 2 lab RNAseq Nicotiana attenuata 2 lab Microarray Volkers et al. 2013 von Reumont et al. 2014 von Schalburg et al. 2008 Caenorhabditis elegans 1, 3 lab Microarray Speleonectes tulumensis 3 field RNAseq Oncorhynchus mykiss 1 field Microarray Vonk et al. 2013 Ophiophagus hannah 1 lab RNAseq Voolstra et al. 2009 multiple 1, 2 lab Microarray Vornanen et al. 2005 Oncorhynchus mykiss 2 lab Microarray Van de Waal et al. 2013 van Ommen Kloeke et al. 2012 Van Puyvelde et al. 2011 Vellichirammal et al. 2014 15 Wägele et al. 2011 multiple 1 lab RNAseq Wang et al. 2013 Chlamys farreri 1 lab RNAseq Wang, H. et al. 2012 Locusta migratoria 1, 2 lab Microarray Wang, M et al. 2011 Anopheles gambiae 2 lab Microarray Wang, S. et al. 2011 Metarhizium robertsii 1, 3 lab Microarray Wang, X. et al. 2011 Bemisia tabaci 1 lab RNAseq Wang, X. et al. 2012 Bemisia tabaci 1 lab RNAseq Wang, X. et al. 2014 Locusta migratoria 1 lab RNAseq Wang, Z. et al. 2014 Neurospora crassa 2 lab Microarray Warr et al. 2007 Anopheles gambiae 1 lab Microarray Wayne et al. 2010 Drosophila spp. 2 lab Microarray Webster et al. 2011 Lagopus lagopus scoticus 2 field Microarray Wei, D. et al. 2013 Liposcelis entomophila 1 lab RNAseq Wei, X. et al. 2013 Barbarea vulgaris 2 lab RNAseq Weiss et al. 2013 Wellband and Heath 2013 Acropora millepora 1 lab RNAseq Oncorhynchus mykiss 1, 2 lab Microarray Wenzel et al. 2013 Lagopus lagopus 2 field Microarray Werner et al. 2013 Patella vulgata 1 lab RNAseq Wertheim et al. 2011 Drosophila melanogaster 2 lab Microarray Wheat et al. 2011 Melitaea cinxia 1, 2 lab Microarray Whitehead et al. 2010 Fundulus heteroclitus 1, 2 lab Microarray Whitehead et al. 2011 Fundulus heteroclitus 2 both Microarray Whitehead et al. 2011b Fundulus heteroclitus 1, 2 both Microarray Whitehead et al. 2012 Fundulus spp. 2 field Microarray Whitehead et al. 2012b Fundulus heteroclitus 2 lab Microarray Whitehead et al. 2012c Fundulus heteroclitus 2, 3 lab Microarray Whitehead et al. 2013 Fundulus spp. 1, 2, 3 lab Microarray Whiteley et al. 2008 Coregonus spp. 1, 3 lab Microarray Whiteman et al. 2012 Whittle and Johannesson 2013 Scaptomyza flava 1, 3 lab RNAseq 1 lab Microarray Wier et al. 2010 Neurospora crassa Euprymna scolopes, Vibrio fischeri 1 field Microarray Williams et al. 2008 Platichthys flesus 2 lab Microarray 16 Wisecaver and Hackett 2010 multiple 1 lab RNAseq Wolf et al. 2010 Corvus corone 1 field RNAseq Wong et al. 2013 Tachyglossus aculeatus 3 lab RNAseq Woodard et al. 2011 multiple 1 field RNAseq Wurmsur et al. 2011 Drosophila spp. 1 lab Microarray Xi et al. 2008 Drosophila melanogaster 2, 3 lab Microarray Xiang et al. 2010 Lateolabrax japonicus 2 lab RNAseq Xu and James 2012 Megachile rotundata 2, 3 lab Microarray Xu et al. 2012 Sogatella furcifera 2 lab RNAseq Xu et al. 2013 Leuciscus waleckii 1, 2 field RNAseq Xue et al. 2010 Nilaparvata lugens 1 lab RNAseq Yampolsky et al. 2012 Drosophila melanogaster 2 lab Microarray Yanai et al. 2011 Xenopus spp. 1 lab Microarray Yang et al. 2011 Alexandrium minutum 2 lab Microarray Yang et al. 2012 Rana spp. 1 field RNAseq Yek et al. 2013 Acromyrmex echinatior 2 lab RNAseq Ying et al. 2013 Escherichia coli 2 lab Microarray Yoo et al. 2010 Yoshida and Ogura 2011 Nuphar advena 1 field Microarray Idiosepius paradoxus 1 lab Microarray Yuan et al. 2012 Deinococcus gobiensis 2 lab RNAseq Yuan et al. 2013 Litopenaeus vannamei 1 lab RNAseq Yudkovski et al. 2010 Lithognathus mormyrus 1, 2 field Microarray Zahn et al. 2010 Eschscholzia californica 1 lab Microarray Zakas et al. 2012 Streblospio benedicti 1 lab RNAseq Zhai et al. 2013 Nilaparvata lugens 1 lab RNAseq Zhang et al. 2006 Methanosarcina barkeri 2 lab Microarray Zhang et al. 2011 Xiphophorus maculatus 1 lab RNAseq Zhang et al. 2012 Arabidopsis thaliana 2 lab Microarray Zhang, L. et al. 2013 Primula spp. 1 field RNAseq Zhang, W. et al. 2013 Papilio spp. 1 both RNAseq Zhang, Y. et al. 2013 Sesamia inferens 1 lab RNAseq Zhao, S. et al. 2013 Collodictyon spp. 1 lab RNAseq 17 Zhao, Z. et al. 2013 Helicoverpa armigera 2 lab RNAseq Zheng et al. 2012 Bactrocera dorsalis 1 lab RNAseq Zheng et al. 2013 Nannochloropsis spp. 1 lab RNAseq Zhou et al. 2013 Desulfovibrio vulgaris 2, 3 lab Microarray Zhou et al. 2014 Apostichopus japonicus 1 lab RNAseq Zhu et al. 2012 Tomicus yunnanensis 1 field RNAseq Zhu et al. 2013 Rhyacionia leptotubula 1 field RNAseq Zhu et al. 2014 Acipenser baerii 1 lab RNAseq REFERENCES Abu‐ Abied, M., Szwerdszarf, D., Mordehaev, I., Levy, A., Belausov, E., Yaniv, Y., . . . Ophir, R. (2012). Microarray analysis revealed upregulation of nitrate reductase in juvenile cuttings of Eucalyptus grandis, which correlated with increased nitric oxide production and adventitious root formation. The Plant Journal, 71(5), 787-799. Aguilar, R., Simard, F., Kamdem, C., Shields, T., Glass, G. E., Garver, L. S., & Dimopoulos, G. (2010). Genome-wide analysis of transcriptomic divergence between laboratory colony and field Anopheles gambiae mosquitoes of the M and S molecular forms. Insect Molecular Biology, 19(5), 695-705. doi: 10.1111/j.1365-2583.2010.01031.x Anderson, J. M., Oliveira, F., Kamhawi, S., Mans, B. J., Reynoso, D., Seitz, A. E., . . . Valenzuela, J. G. (2006). Comparative salivary gland transcriptomics of sandfly vectors of visceral leishmaniasis. BMC genomics, 7. doi: 10.1186/1471-2164-7-52 Andersson, M. N., Grosse-Wilde, E., Keeling, C. I., Bengtsson, J. M., Yuen, M. M. S., Li, M., . . . Schlyter, F. (2013). Antennal transcriptome analysis of the chemosensory gene families in the tree killing bark beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae). BMC genomics, 14. doi: 10.1186/1471-2164-14-198 Andres, J. A., Larson, E. L., Bogdanowicz, S. M., & Harrison, R. G. (2013). Patterns of Transcriptome Divergence in the Male Accessory Gland of Two Closely Related Species of Field Crickets. Genetics, 193(2), 501-+. doi: 10.1534/genetics.112.142299 Aranda, M., Banaszak, A. T., Bayer, T., Luyten, J. R., Medina, M., & Voolstra, C. R. (2011). Differential sensitivity of coral larvae to natural levels of ultraviolet radiation during the onset of larval competence. Molecular Ecology, 20(14), 2955-2972. doi: 10.1111/j.1365-294X.2011.05153.x Arca, B., Lombardo, F., Valenzuela, J. G., Francischetti, I. M. B., Marinotti, O., Coluzzi, M., & Ribeiro, J. A. C. (2005). An updated catalogue of salivary gland transcripts in the adult female mosquito, Anopheles gambiae. Journal of Experimental Biology, 208(20), 3971-3986. doi: 10.1242/jcb.01849 Arfi, Y., Levasseur, A., & Record, E. (2013). Differential Gene Expression in Pycnoporus coccineus during Interspecific Mycelial Interactions with Different Competitors. Applied and Environmental Microbiology, 79(21), 6626-6636. doi: 10.1128/aem.02316-13 Arnaiz, O., Gout, J.-F., Betermier, M., Bouhouche, K., Cohen, J., Duret, L., . . . Sperling, L. (2010). Gene expression in a paleopolyploid: a transcriptome resource for the ciliate Paramecium tetraurelia. BMC genomics, 11. doi: 10.1186/1471-2164-11-547 Aubin-Horth, N., Letcher, B. H., & Hofmann, H. A. (2009). Gene-expression signatures of Atlantic salmon's plastic life cycle. Gen Comp Endocrinol, 163(3), 278-284. doi: 10.1016/j.ygcen.2009.04.021 18 Avarre, J. C., Dugue, R., Alonso, P., Diombokho, A., Joffrois, C., Faivre, N., . . . Durand, J. D. (2014). Analysis of the black-chinned tilapia Sarotherodon melanotheron heudelotii reproducing under a wide range of salinities: from RNA-seq to candidate genes. Mol Ecol Resour, 14(1), 139-149. doi: 10.1111/1755-0998.12148 Babik, W., Stuglik, M., Qi, W., Kuenzli, M., Kuduk, K., Koteja, P., & Radwan, J. (2010). Heart transcriptome of the bank vole (Myodes glareolus): towards understanding the evolutionary variation in metabolic rate. BMC genomics, 11. doi: 10.1186/1471-2164-11-390 Bachmann, H., Starrenburg, M. J. C., Molenaar, D., Kleerebezem, M., & Vlieg, J. E. T. v. H. (2012). Microbial domestication signatures of Lactococcus lactis can be reproduced by experimental evolution. Genome Research, 22(1), 115-124. doi: 10.1101/gr.121285.111 Bachvaroff, T. R., Gornik, S. G., Concepcion, G. T., Waller, R. F., Mendez, G. S., Lippmeier, J. C., & Delwiche, C. F. (2014). Dinoflagellate phylogeny revisited: Using ribosomal proteins to resolve deep branching dinoflagellate clades. Molecular phylogenetics and evolution, 70, 314-322. doi: 10.1016/j.ympev.2013.10.007 Badisco, L., Huybrechts, J., Simonet, G., Verlinden, H., Marchal, E., Huybrechts, R., . . . Broeck, J. V. (2011). Transcriptome analysis of the desert locust central nervous system: production and annotation of a Schistocerca gregaria EST database. PloS one, 6(3), e17274. Badouin, H., Belkhir, K., Gregson, E., Galindo, J., Sundstrom, L., Martin, S. J., . . . Smadja, C. M. (2013). Transcriptome Characterisation of the Ant Formica exsecta with New Insights into the Evolution of Desaturase Genes in Social Hymenoptera. PloS one, 8(7). doi: 10.1371/journal.pone.0068200 Bai, Z., Zheng, H., Lin, J., Wang, G., & Li, J. (2013). Comparative analysis of the transcriptome in tissues secreting purple and white nacre in the pearl mussel Hyriopsis cumingii. PloS one, 8(1), e53617. Bai, X., Zhang, W., Orantes, L., Jun, T.-H., Mittapalli, O., Mian, M. A. R., & Michel, A. P. (2010). Combining Next-Generation Sequencing Strategies for Rapid Molecular Resource Development from an Invasive Aphid Species, Aphis glycines. PloS one, 5(6). doi: 10.1371/journal.pone.0011370 Bajgain, P., Richardson, B. A., Price, J. C., Cronn, R. C., & Udall, J. A. (2011). Transcriptome characterization and polymorphism detection between subspecies of big sagebrush (Artemisia tridentata). BMC genomics, 12. doi: 10.1186/1471-2164-12-370 Balakrishnan, C. N., Chapus, C., Brewer, M. S., & Clayton, D. F. (2013). Brain transcriptome of the violet-eared waxbill Uraeginthus granatina and recent evolution in the songbird genome. Open Biology, 3(9). doi: 10.1098/rsob.130063 Baldo, L., Santos, M. E., & Salzburger, W. (2011). Comparative Transcriptomics of Eastern African Cichlid Fishes Shows Signs of Positive Selection and a Large Contribution of Untranslated Regions to Genetic Diversity. Genome Biology and Evolution, 3, 443-455. doi: 10.1093/gbe/evr047 Ballerini, E. S., Mockaitis, K., & Arnold, M. L. (2013). Transcriptome sequencing and phylogenetic analysis of floral and leaf MIKCC MADS-box and R2R3 MYB transcription factors from the monocot Iris fulva. Gene, 531(2), 337-346. doi: 10.1016/j.gene.2013.08.067 Banni, M., Negri, A., Mignone, F., Boussetta, H., Viarengo, A., & Dondero, F. (2011). Gene Expression Rhythms in the Mussel Mytilus galloprovincialis (Lam.) across an Annual Cycle. PloS one, 6(5). doi: 10.1371/journal.pone.0018904 Bao, Y.-Y., Qu, L.-Y., Zhao, D., Chen, L.-B., Jin, H.-Y., Xu, L.-M., . . . Zhang, C.-X. (2013). The genome- and transcriptome-wide analysis of innate immunity in the brown planthopper, Nilaparvata lugens. BMC genomics, 14. doi: 10.1186/1471-2164-14-160 Bao, Y.-Y., Wang, Y., Wu, W.-J., Zhao, D., Xue, J., Zhang, B.-Q., . . . Zhang, C.-X. (2012). De novo intestine-specific transcriptome of the brown planthopper Nilaparvata lugens revealed potential functions in digestion, detoxification and immune response. Genomics, 99(4), 256-264. doi: 10.1016/j.ygeno.2012.02.002 19 Bar-Yaacov, D., Bouskila, A., & Mishmar, D. (2013). The First Chameleon Transcriptome: Comparative Genomic Analysis of the OXPHOS System Reveals Loss of COX8 in Iguanian Lizards. Genome Biology and Evolution, 5(10), 1792-1799. doi: 10.1093/gbe/evt131 Barah, P., Winge, P., Kusnierczyk, A., Tran, D. H., & Bones, A. M. (2013). Molecular Signatures in Arabidopsis thaliana in Response to Insect Attack and Bacterial Infection. PloS one, 8(3). doi: 10.1371/journal.pone.0058987 Bardil, A., de Almeida, J. D., Combes, M. C., Lashermes, P., & Bertrand, B. (2011). Genomic expression dominance in the natural allopolyploid Coffea arabica is massively affected by growth temperature. New Phytologist, 192(3), 760-774. doi: 10.1111/j.1469-8137.2011.03833.x Barreto, F. S., Moy, G. W., & Burton, R. S. (2011). Interpopulation patterns of divergence and selection across the transcriptome of the copepod Tigriopus californicus. Molecular Ecology, 20(3), 560-572. doi: 10.1111/j.1365-294X.2010.04963.x Barshis, D. J., Ladner, J. T., Oliver, T. A., Seneca, F. O., Traylor-Knowles, N., & Palumbi, S. R. (2013). Genomic basis for coral resilience to climate change. Proc Natl Acad Sci U S A, 110(4), 1387-1392. doi: 10.1073/pnas.1210224110 Bass, C., Hebsgaard, M. B., & Hughes, J. (2012). Genomic resources for the brown planthopper, Nilaparvata lugens: Transcriptome pyrosequencing and microarray design. Insect Science, 19(1), 1-12. doi: 10.1111/j.1744-7917.2011.01440.x Bassene, J. B., Froelicher, Y., Dubois, C., Ferrer, R. M., Navarro, L., Ollitrault, P., & Ancillo, G. (2010). Non-additive gene regulation in a citrus allotetraploid somatic hybrid between C. reticulata Blanco and C. limon (L.) Burm. Heredity, 105(3), 299-308. doi: 10.1038/hdy.2009.162 Baumgarten, S., Bayer, T., Aranda, M., Liew, Y. J., Carr, A., Micklem, G., & Voolstra, C. R. (2013). Integrating microRNA and mRNA expression profiling in Symbiodinium microadriaticum, a dinoflagellate symbiont of reef-building corals. BMC genomics, 14, 1-18. Bay, L. K., Nielsen, H. B., Jarmer, H., Seneca, F., & van Oppen, M. J. (2009). Transcriptomic variation in a coral reveals pathways of clonal organisation. Mar Genomics, 2(2), 119-125. Bay, L. K., Ulstrup, K. E., Nielsen, H. B., Jarmer, H., Goffard, N., Willis, B. L., . . . Van Oppen, M. J. (2009). Microarray analysis reveals transcriptional plasticity in the reef building coral Acropora millepora. Mol Ecol, 18(14), 3062-3075. doi: 10.1111/j.1365-294X.2009.04257.x Bayne, C. J., Gerwick, L., Wheeler, P. A., & Thorgaard, G. H. (2006). Transcriptome profiles of livers and kidneys from three rainbow trout (Oncorhynchus mykiss) clonal lines distinguish stocks from three allopatric populations. Comparative Biochemistry and Physiology D-Genomics & Proteomics, 1(4), 396-403. doi: 10.1016/j.cbd.2006.10.001 Bazinet, A. L., Cummings, M. P., Mitter, K. T., & Mitter, C. W. (2013). Can RNA-Seq Resolve the Rapid Radiation of Advanced Moths and Butterflies (Hexapoda: Lepidoptera: Apoditrysia)? An Exploratory Study. PloS one, 8(12). doi: 10.1371/journal.pone.0082615 Becks, L., Ellner, S. P., Jones, L. E., & Hairston, N. G. (2012). The functional genomics of an eco‐ evolutionary feedback loop: linking gene expression, trait evolution, and community dynamics. Ecology Letters, 15(5), 492-501. Beemster, G. T. S., Vercruysse, S., De Veylder, L., Kuiper, M., & Inze, D. (2006). The Arabidopsis leaf as a model system for investigating the role of cell cycle regulation in organ growth. Journal of Plant Research, 119(1), 43-50. doi: 10.1007/s10265-005-0234-2 Belgard, T. G., Montiel, J. F., Wang, W. Z., Garcia-Moreno, F., Margulies, E. H., Ponting, C. P., & Molnar, Z. (2013). Adult pallium transcriptomes surprise in not reflecting predicted homologies across diverse chicken and mouse pallial sectors. Proc Natl Acad Sci U S A, 110(32), 13150-13155. doi: 10.1073/pnas.1307444110 20 Bermingham, J., Rabatel, A., Calevro, F., Vinuelas, J., Febvay, G., Charles, H., . . . Wilkinson, T. (2009). Impact of Host Developmental Age on the Transcriptome of the Symbiotic Bacterium Buchnera aphidicola in the Pea Aphid (Acyrthosiphon pisum). Applied and Environmental Microbiology, 75(22), 7294-7297. doi: 10.1128/aem.01472-09 Bernatchez, L., St-Cyr, J., Normandeau, E., Maes, G. E., Als, T. D., Kalujnaia, S., . . . Hansen, M. M. (2011). Differential timing of gene expression regulation between leptocephali of the two Anguilla eel species in the Sargasso Sea. Ecology and Evolution, 1(4). doi: 10.1002/ece3.27 Bettencourt, R., Pinheiro, M., Egas, C., Gomes, P., Afonso, M., Shank, T., & Santos, R. S. (2010). Highthroughput sequencing and analysis of the gill tissue transcriptome from the deep-sea hydrothermal vent mussel Bathymodiolus azoricus. BMC genomics, 11. doi: 10.1186/1471-2164-11-559 Bilyk, K. T., & Cheng, C. H. C. (2013). Model of gene expression in extreme cold - reference transcriptome for the high-Antarctic cryopelagic notothenioid fish Pagothenia borchgrevinki. BMC genomics, 14, 1-16. Boltana, S., Rey, S., Roher, N., Vargas, R., Huerta, M., Huntingford, F. A., . . . MacKenzie, S. (2013). Behavioural fever is a synergic signal amplifying the innate immune response. Proceedings of the Royal Society Biological Sciences Series B, 280(1766), 1-11. Bonneau, L., Huguet, S., Wipf, D., Pauly, N., & Hoai-Nam, T. (2013). Combined phosphate and nitrogen limitation generates a nutrient stress transcriptome favorable for arbuscular mycorrhizal symbiosis in Medicago truncatula. New Phytologist, 199(1), 188-202. doi: 10.1111/nph.12234 Bonneaud, C., Balenger, S. L., Russell, A. F., Zhang, J., Hill, G. E., & Edwards, S. V. (2011). Rapid evolution of disease resistance is accompanied by functional changes in gene expression in a wild bird. Proceedings of the National Academy of Sciences, 108(19), 7866-7871. Bougas, B., Audet, C., & Bernatchez, L. (2013). The influence of parental effects on transcriptomic landscape during early development in brook charr (Salvelinus fontinalis, Mitchill). Heredity, 110(5), 484-491. doi: 10.1038/hdy.2012.113 Bougas, B., Granier, S., Audet, C., & Bernatchez, L. (2010). The Transcriptional Landscape of CrossSpecific Hybrids and Its Possible Link With Growth in Brook Charr (Salvelinus fontinalis Mitchill). Genetics, 186(1), 97-U207. doi: 10.1534/genetics.110.118158 Boulet, M., Normandeau, E., Bougas, B., Audet, C., & Bernatchez, L. (2012). Comparative transcriptomics of anadromous and resident brook charr Salvelinus fontinalis before their first salt water transition. Current Zoology, 58(1), 158-170. Brady, A. K., Snyder, K. A., & Vize, P. D. (2011). Circadian Cycles of Gene Expression in the Coral, Acropora millepora. PloS one, 6(9). doi: 10.1371/journal.pone.0025072 Brawand, D., Soumillon, M., Necsulea, A., Julien, P., Csárdi, G., Harrigan, P., . . . Kircher, M. (2011). The evolution of gene expression levels in mammalian organs. Nature, 478(7369), 343-348. Briscoe, A. D., Macias-Munoz, A., Kozak, K. M., Walters, J. R., Yuan, F., Jamie, G. A., . . . Jiggins, C. D. (2013). Female Behaviour Drives Expression and Evolution of Gustatory Receptors in Butterflies. PLoS genetics, 9(7), 1-16. Broekgaarden, C., Poelman, E. H., Steenhuis, G., Voorrips, R. E., Dicke, M., & Vosman, B. (2008). Responses of Brassica oleracea cultivars to infestation by the aphid Brevicoryne brassicae: an ecological and molecular approach. Plant Cell and Environment, 31(11), 1592-1605. doi: 10.1111/j.1365-3040.2008.01871.x Broekgaarden, C., Voorrips, R. E., Dicke, M., & Vosman, B. (2011). Transcriptional responses of Brassica nigra to feeding by specialist insects of different feeding guilds. Insect Science, 18(3), 259272. doi: 10.1111/j.1744-7917.2010.01368.x Bruce, T. J. A., Matthes, M. C., Chamberlain, K., Woodcock, C. M., Mohib, A., Webster, B., . . . Napier, J. A. (2008). cis-Jasmone induces Arabidopsis genes that affect the chemical ecology of multitrophic 21 interactions with aphids and their parasitoids. Proc Natl Acad Sci U S A, 105(12), 4553-4558. doi: 10.1073/pnas.0710305105 Bryk, J., Somel, M., Lorenc, A., & Teschke, M. (2013). Early gene expression divergence between allopatric populations of the house mouse (Mus musculus domesticus). Ecology and Evolution, 3(3), 558-568. Buckley, B. A., Gracey, A. Y., & Somero, G. N. (2006). The cellular response to heat stress in the goby Gillichthys mirabilis: a cDNA microarray and protein-level analysis. Journal of Experimental Biology, 209(14), 2660-2677. doi: 10.1242/jeb.02292 Burke, G. R., & Moran, N. A. (2011). Responses of the pea aphid transcriptome to infection by facultative symbionts. Insect Molecular Biology, 20(3), 357-365. doi: 10.1111/j.1365-2583.2011.01070.x Burke, G. R., & Strand, M. R. (2014). Systematic analysis of a wasp parasitism arsenal. Molecular Ecology, 23(4), 890-+. doi: 10.1111/mec.12648 Burki, F., Corradi, N., Sierra, R., Pawlowski, J., Meyer, G. R., Abbott, C. L., & Keeling, P. J. (2013). Phylogenomics of the Intracellular Parasite Mikrocytos mackini Reveals Evidence for a Mitosome in Rhizaria. Current Biology, 23(16), 1541-1547. doi: 10.1016/j.cub.2013.06.033 Burns, G., Ortega-Martinez, O., Dupont, S., Thorndyke, M. C., Peck, L. S., & Clark, M. S. (2012). Intrinsic gene expression during regeneration in arm explants of Amphiura filiformis. Journal of Experimental Marine Biology and Ecology, 413, 106-112. doi: 10.1016/j.jembe.2011.12.001 Burns, G., Thorndyke, M. C., Peck, L. S., & Clark, M. S. (2013). Transcriptome pyrosequencing of the Antarctic brittle star Ophionotus victoriae. Mar Genomics, 9, 9-15. doi: 10.1016/j.margen.2012.05.003 Busby, M. A., Gray, J. M., Costa, A. M., Stewart, C., Stromberg, M. P., Barnett, D., . . . Marth, G. T. (2011). Expression divergence measured by transcriptome sequencing of four yeast species. BMC genomics, 12. doi: 10.1186/1471-2164-12-635 Calla, B., Hall, B., Hou, S., & Geib, S. M. (2014). A genomic perspective to assessing quality of massreared SIT flies used in Mediterranean fruit fly (Ceratitis capitata) eradication in California. BMC genomics, 15. doi: 10.1186/1471-2164-15-98 Campanaro, S., Vezzi, A., Vitulo, N., Lauro, F. M., D'Angelo, M., Simonato, F., . . . Bartlett, D. H. (2005). Laterally transferred elements and high pressure adaptation in Photobacterium profundum strains. BMC genomics, 6. doi: 10.1186/1471-2164-6-122 Cardoen, D., Wenseleers, T., Ernst, U. R., Danneels, E. L., Laget, D., De Graaf, D. C., . . . Verleyen, P. (2011). Genome‐ wide analysis of alternative reproductive phenotypes in honeybee workers. Molecular Ecology, 20(19), 4070-4084. Carvalhais, L. C., Dennis, P. G., Fan, B., Fedoseyenko, D., Kierul, K., Becker, A., ... & Borriss, R. (2013). Linking Plant Nutritional Status to Plant-Microbe Interactions. PloS one, 8(7), e68555. Cassone, B. J., Molloy, M. J., Cheng, C., Tan, J. C., Hahn, M. W., & Besansky, N. J. (2011). Divergent transcriptional response to thermal stress by Anopheles gambiae larvae carrying alternative arrangements of inversion 2La. Molecular Ecology, 20(12), 2567-2580. doi: 10.1111/j.1365294X.2011.05114.x Castilho, P. C., Buckley, B. A., Somero, G., & Block, B. A. (2009). Heterologous hybridization to a complementary DNA microarray reveals the effect of thermal acclimation in the endothermic bluefin tuna (Thunnus orientalis). Molecular Ecology, 18(10), 2092-2102. doi: 10.1111/j.1365294X.2009.04174.x Castoe, T. A., de Koning, A. P. J., Hall, K. T., Card, D. C., Schield, D. R., Fujita, M. K., . . . Pollock, D. D. (2013). The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc Natl Acad Sci U S A, 110(51), 20645-20650. doi: 10.1073/pnas.1314475110 22 Celorio-Mancera, M. d. l. P., Wheat, C. W., Vogel, H., Soderlind, L., Janz, N., & Nylin, S. (2013). Mechanisms of macroevolution: polyphagous plasticity in butterfly larvae revealed by RNA-Seq. Molecular Ecology, 22(19), 4884-4895. doi: 10.1111/mec.12440 Chan, Z., Bigelow, P. J., Loescher, W., & Grumet, R. (2012). Comparison of salt stress resistance genes in transgenic Arabidopsis thaliana indicates that extent of transcriptomic change may not predict secondary phenotypic or fitness effects. Plant Biotechnol J, 10(3), 284-300. doi: 10.1111/j.14677652.2011.00661.x Chaney, M. L., & Gracey, A. Y. (2011). Mass mortality in Pacific oysters is associated with a specific gene expression signature. Molecular Ecology, 20(14), 2942-2954. doi: 10.1111/j.1365294X.2011.05152.x Chapman, M. A., Hiscock, S. J., & Filatov, D. A. (2013). Genomic Divergence during Speciation Driven by Adaptation to Altitude. Molecular biology and evolution, 30(12), 2553-2567. doi: 10.1093/molbev/mst168 Chapman, R. W., Mancia, A., Beal, M., Veloso, A., Rathburn, C., Blair, A., . . . Sanger, D. (2011). The transcriptomic responses of the eastern oyster, Crassostrea virginica, to environmental conditions. Molecular Ecology, 20(7), 1431-1449. doi: 10.1111/j.1365-294X.2011.05018.x Chapman, R. W., Mancia, A., Beal, M., Veloso, A., Rathburn, C., Blair, A., . . . Didonato, G. (2009). A transcriptomic analysis of land-use impacts on the oyster, Crassostrea virginica, in the South Atlantic bight. Mol Ecol, 18(11), 2415-2425. doi: 10.1111/j.1365-294X.2009.04194.x Chaudhary, B., Hovav, R., Rapp, R., Verma, N., Udall, J. A., & Wendel, J. F. (2008). Global analysis of gene expression in cotton fibers from wild and domesticated Gossypium barbadense. Evolution & Development, 10(5), 567-582. doi: 10.1111/j.1525-142X.2008.00272.x Chelaifa, H., Mahe, F., & Ainouche, M. (2010). Transcriptome divergence between the hexaploid saltmarsh sister species Spartina maritima and Spartina alterniflora (Poaceae). Molecular Ecology, 19(10), 2050-2063. doi: 10.1111/j.1365-294X.2010.04637.x Chelaifa, H., Monnier, A., & Ainouche, M. (2010). Transcriptomic changes following recent natural hybridization and allopolyploidy in the salt marsh species Spartina x townsendii and Spartina anglica (Poaceae). New Phytologist, 186(1), 161-174. doi: 10.1111/j.1469-8137.2010.03179.x Chen, F., Fasoli, M., Tornielli, G. B., Dal Santo, S., Pezzotti, M., Zhang, L., . . . Cheng, Z.-M. (2013). The Evolutionary History and Diverse Physiological Roles of the Grapevine Calcium-Dependent Protein Kinase Gene Family. PloS one, 8(12). doi: 10.1371/journal.pone.0080818 Chen, M., Zou, M., Yang, L., & He, S. (2012). Basal Jawed Vertebrate Phylogenomics Using Transcriptomic Data from Solexa Sequencing. PloS one, 7(4). doi: 10.1371/journal.pone.0036256 Chen, S., Yang, P., Jiang, F., Wei, Y., Ma, Z., & Kang, L. (2010). De Novo Analysis of Transcriptome Dynamics in the Migratory Locust during the Development of Phase Traits. PloS one, 5(12). doi: 10.1371/journal.pone.0015633 Chen, Z.-F., Matsumura, K., Wang, H., Arellano, S. M., Yan, X., Alam, I., . . . Qian, P.-Y. (2011). Toward an Understanding of the Molecular Mechanisms of Barnacle Larval Settlement: A Comparative Transcriptomic Approach. PloS one, 6(7). doi: 10.1371/journal.pone.0022913 Chen, Z., Yu, H., Li, L., Hu, S., & Dong, X. (2012). The genome and transcriptome of a newly described psychrophilic archaeon, Methanolobus psychrophilus R15, reveal its cold adaptive characteristics. Environmental Microbiology Reports, 4(6), 633-641. doi: 10.1111/j.1758-2229.2012.00389.x Cheviron, Z. A., Connaty, A. D., McClelland, G. B., & Storz, J. F. (2014). FUNCTIONAL GENOMICS OF ADAPTATION TO HYPOXIC COLD-STRESS IN HIGH-ALTITUDE DEER MICE: TRANSCRIPTOMIC PLASTICITY AND THERMOGENIC PERFORMANCE. Evolution, 68(1), 48-62. doi: 10.1111/evo.12257 23 Cheviron, Z. A., Whitehead, A., & Brumfield, R. T. (2008). Transcriptomic variation and plasticity in rufous-collared sparrows (Zonotrichia capensis) along an altitudinal gradient. Molecular Ecology, 17(20), 4556-4569. doi: 10.1111/j.1365-294X.2008.03942.x Chiari, Y., Cahais, V., Galtier, N., & Delsuc, F. (2012). Phylogenomic analyses support the position of turtles as the sister group of birds and crocodiles (Archosauria). BMC Biol, 10. doi: 10.1186/17417007-10-65 Chu, J.-H., Lin, R.-C., Yeh, C.-F., Hsu, Y.-C., & Li, S.-H. (2012). Characterization of the transcriptome of an ecologically important avian species, the Vinous-throated Parrotbill Paradoxornis webbianus bulomachus (Paradoxornithidae; Aves). BMC genomics, 13. doi: 10.1186/1471-2164-13-149 Chun, C. K., Troll, J. V., Koroleva, I., Brown, B., Manzella, L., Snir, E., . . . McFall-Ngai, M. J. (2008). Effects of colonization, luminescence, and autoinducer on host transcription during development of the squid-vibrio association. Proc Natl Acad Sci U S A, 105(32), 11323-11328. doi: 10.1073/pnas.0802369105 Clark, M. E., He, Z., Redding, A. M., Joachimiak, M. P., Keasling, J. D., Zhou, J. Z., . . . Fields, M. W. (2012). Transcriptomic and proteomic analyses of Desulfovibrio vulgaris biofilms: Carbon and energy flow contribute to the distinct biofilm growth state. BMC genomics, 13. doi: 10.1186/1471-2164-13138 Clark, M. S., Thorne, M. A., Vieira, F. A., Cardoso, J. C., Power, D. M., & Peck, L. S. (2010). Insights into shell deposition in the Antarctic bivalve Laternula elliptica: gene discovery in the mantle transcriptome using 454 pyrosequencing. BMC genomics, 11(1), 362. Coate, J. E., & Doyle, J. J. (2010). Quantifying Whole Transcriptome Size, a Prerequisite for Understanding Transcriptome Evolution Across Species: An Example from a Plant Allopolyploid. Genome Biology and Evolution, 2, 534-546. doi: 10.1093/gbe/evq038 Coate, J. E., Powell, A. F., Owens, T. G., & Doyle, J. J. (2013). Transgressive physiological and transcriptomic responses to light stress in allopolyploid Glycine dolichocarpa (Leguminosae). Heredity, 110(2), 160-170. doi: 10.1038/hdy.2012.77 Cohen, R., Chalifa-Caspi, V., Williams, T. D., Auslander, M., George, S. G., Chipman, J. K., & Tom, M. (2007). Estimating the efficiency of fish cross-species cDNA microarray hybridization. Marine Biotechnology, 9(4), 491-499. doi: 10.1007/s10126-007-9010-8 Collen, J., Guisle-Marsollier, I., Leger, J. J., & Boyen, C. (2007). Response of the transcriptome of the intertidal red seaweed Chondrus crispus to controlled and natural stresses. New Phytologist, 176(1), 45-55. doi: 10.1111/j.1469-8137.2007.02152.x Colombo, M., Diepeveen, E. T., Muschick, M., Santos, M. E., Indermaur, A., Boileau, N., . . . Salzburger, W. (2013). The ecological and genetic basis of convergent thick-lipped phenotypes in cichlid fishes. Molecular Ecology, 22(3), 670-684. doi: 10.1111/mec.12029 Comeault, A. A., Sommers, M., Schwander, T., Buerkle, C. A., Farkas, T. E., Nosil, P., & Parchman, T. L. (2012). De novo characterization of the Timema cristinae transcriptome facilitates marker discovery and inference of genetic divergence. Mol Ecol Resour, 12(3), 549-561. doi: 10.1111/j.17550998.2012.03121.x Conaco, C., Neveu, P., Zhou, H., Arcila, M. L., Degnan, S. M., Degnan, B. M., & Kosik, K. S. (2012). Transcriptome profiling of the demosponge Amphimedon queenslandica reveals genome-wide events that accompany major life cycle transitions. BMC genomics, 13. doi: 10.1186/1471-2164-13-209 Conant, G. C., & Wagner, A. (2004). Duplicate genes and robustness to transient gene knock-downs in Caenorhabditis elegans. Proceedings of the Royal Society of London. Series B: Biological Sciences, 271(1534), 89-96. Connor, K. M., & Gracey, A. Y. (2011). Circadian cycles are the dominant transcriptional rhythm in the intertidal mussel Mytilus californianus. Proc Natl Acad Sci U S A, 108(38), 16110-16115. doi: 10.1073/pnas.1111076108 24 Cook, N., Aziz, N., Hedley, P. E., Morris, J., Milne, L., Karley, A. J., . . . Russell, J. R. (2011). Transcriptome sequencing of an ecologically important graminivorous sawfly: a resource for marker development. Conservation Genetics Resources, 3(4), 789-795. doi: 10.1007/s12686-011-9459-7 Coppe, A., Pujolar, J. M., Maes, G. E., Larsen, P. F., Hansen, M. M., Bernatchez, L., . . . Bortoluzzi, S. (2010). Sequencing, de novo annotation and analysis of the first Anguilla anguilla transcriptome: EeelBase opens new perspectives for the study of the critically endangered european eel. BMC genomics, 11. doi: 10.1186/1471-2164-11-635 Corredor-Adamez, M., Welten, M. C. M., Spaink, H. P., Jeffery, J. E., Schoon, R. T., de Bakker, M. A. G., . . . Richardson, M. K. (2005). Genomic annotation and transcriptome analysis of the zebrafish (Danio rerio) hox complex with description of a novel member, hoxb13a. Evolution & Development, 7(5), 362-375. doi: 10.1111/j.1525-142X.2005.05042.x Cseke, L. J., Tsai, C.-J., Rogers, A., Nelsen, M. P., White, H. L., Karnosky, D. F., & Podila, G. K. (2009). Transcriptomic comparison in the leaves of two aspen genotypes having similar carbon assimilation rates but different partitioning patterns under elevated CO2. New Phytologist, 182(4), 891-911. doi: 10.1111/j.1469-8137.2009.02812.x Cui, R., Schumer, M., Kruesi, K., Walter, R., Andolfatto, P., & Rosenthal, G. G. (2013). PHYLOGENOMICS REVEALS EXTENSIVE RETICULATE EVOLUTION IN XIPHOPHORUS FISHES. Evolution, 67(8), 2166-2179. doi: 10.1111/evo.12099 Cullingham, C. I., Cooke, J. E. K., & Coltman, D. W. (2013). Effects of introgression on the genetic population structure of two ecologically and economically important conifer species: lodgepole pine (Pinus contorta var. latifolia) and jack pine (Pinus banksiana). Genome, 56(10), 577-585. doi: 10.1139/gen-2013-0071 Czesny, S., Epifanio, J., & Michalak, P. (2012). Genetic Divergence between Freshwater and Marine Morphs of Alewife (Alosa pseudoharengus): A 'Next-Generation' Sequencing Analysis. PloS one, 7(3). doi: 10.1371/journal.pone.0031803 Czypionka, T., Cheng, J., Pozhitkov, A., & Nolte, A. W. (2012). Transcriptome changes after genomewide admixture in invasive sculpins (Cottus). Molecular Ecology, 21(19), 4797-4810. doi: 10.1111/j.1365-294X.2012.05645.x Davidson, R. M., Gowda, M., Moghe, G., Lin, H., Vaillancourt, B., Shiu, S.-H., . . . Buell, C. R. (2012). Comparative transcriptomics of three Poaceae species reveals patterns of gene expression evolution. Plant Journal, 71(3), 492-502. doi: 10.1111/j.1365-313X.2012.05005.x de Boer, T. E., Birlutiu, A., Bochdanovits, Z., Timmermans, M. J. T. N., Dijkstra, T. M. H., Van Straalen, N. M., . . . Roelofs, D. (2011). Transcriptional plasticity of a soil arthropod across different ecological conditions. Molecular Ecology, 20(6), 1144-1154. doi: 10.1111/j.1365-294X.2010.04985.x de Carvalho, J. F., Poulain, J., Da Silva, C., Wincker, P., Michon-Coudouel, S., Dheilly, A., . . . Ainouche, M. (2013). Transcriptome de novo assembly from next-generation sequencing and comparative analyses in the hexaploid salt marsh species Spartina maritima and Spartina alterniflora (Poaceae). Heredity, 110(2), 181-193. doi: 10.1038/hdy.2012.76 De Wit, P., & Palumbi, S. R. (2013). Transcriptome-wide polymorphisms of red abalone (Haliotis rufescens) reveal patterns of gene flow and local adaptation. Molecular Ecology, 22(11), 2884-2897. doi: 10.1111/mec.12081 Debes, P. V., Normandeau, E., Fraser, D. J., Bernatchez, L., & Hutchings, J. A. (2012). Differences in transcription levels among wild, domesticated, and hybrid Atlantic salmon (Salmo salar) from two environments. Molecular Ecology, 21(11), 2574-2587. Denekamp, N. Y., Reinhardt, R., Albrecht, M. W., Drungowski, M., Kube, M., & Lubzens, E. (2011). The expression pattern of dormancy-associated genes in multiple life-history stages in the rotifer Brachionus plicatilis. Hydrobiologia, 662(1), 51-63. doi: 10.1007/s10750-010-0518-y 25 Denver, D. R., Morris, K., Streelman, J. T., Kim, S. K., Lynch, M., & Thomas, W. K. (2005). The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nature genetics, 37(5), 544-548. doi: 10.1038/ng1554 Der, J. P., Barker, M. S., Wickett, N. J., dePamphilis, C. W., & Wolf, P. G. (2011). De novo characterization of the gametophyte transcriptome in bracken fern, Pteridium aquilinum. BMC genomics, 12. doi: 10.1186/1471-2164-12-99 Derome, N., & Bernatchez, L. (2006). The transcriptomics of ecological convergence between 2 limnetic coregonine fishes (Salmonidae). Molecular biology and evolution, 23(12), 2370-2378. doi: 10.1093/molbev/msl110 Derome, N., Duchesne, P., & Bernatchez, L. (2006). Parallelism in gene transcription among sympatric lake whitefish (Coregonus clupeaformis Mitchill) ecotypes. Molecular Ecology, 15(5), 1239-1249. Derome, N., Bougas, B., Rogers, S. M., Whiteley, A., Labbe, A., Laroche, J., & Bernatchez, L. (2008). Pervasive sex-linked effects on transcription regulation as revealed by expression quantitative trait loci mapping in lake whitefish species pairs (Coregonus sp., Salmonidae). Genetics, 179(4), 19031917. Des Marais, D. L., McKay, J. K., Richards, J. H., Sen, S., Wayne, T., & Juenger, T. E. (2012). Physiological Genomics of Response to Soil Drying in Diverse Arabidopsis Accessions. Plant Cell, 24(3), 893-914. doi: 10.1105/tpc.112.096180 DeSalvo, M. K., Sunagawa, S., Fisher, P. L., Voolstra, C. R., Iglesias-Prieto, R., & Medina, M. (2010). Coral host transcriptomic states are correlated with Symbiodinium genotypes. Molecular Ecology, 19(6), 1174-1186. doi: 10.1111/j.1365-294X.2010.04534.x DeSalvo, M. K., Sunagawa, S., Voolstra, C. R., & Medina, M. (2010). Transcriptomic responses to heat stress and bleaching in the elkhorn coral Acropora palmata. Marine Ecology Progress Series, 402, 97113. Desalvo, M. K., Voolstra, C. R., Sunagawa, S., Schwarz, J. A., Stillman, J. H., Coffroth, M. A., . . . Medina, M. (2008). Differential gene expression during thermal stress and bleaching in the Caribbean coral Montastraea faveolata. Molecular Ecology, 17(17), 3952-3971. doi: 10.1111/j.1365294X.2008.03879.x Dhar, R., Saegesser, R., Weikert, C., Yuan, J., & Wagner, A. (2011). Adaptation of Saccharomyces cerevisiae to saline stress through laboratory evolution. J Evol Biol, 24(5), 1135-1153. doi: 10.1111/j.1420-9101.2011.02249.x Domazet-Loso, T., & Tautz, D. (2010). A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature, 468(7325), 815-U107. doi: 10.1038/nature09632 Doroszuk, A., Jonker, M. J., Pul, N., Breit, T. M., & Zwaan, B. J. (2012). Transcriptome analysis of a long-lived natural Drosophila variant: a prominent role of stress-and reproduction-genes in lifespan extension. BMC genomics, 13(1), 167. dos Santos Ferreira, V., Rocchetta, L., Conforti, V., Bench, S., Feldman, R., & Levin, M. J. (2007). Gene expression patterns in Euglena gracilis: insights into the cellular response to environmental stress. Gene (Amsterdam), 389(2), 136-145. doi: 10.1016/j.gene.2006.10.023 Dou, W., Shen, G.-M., Niu, J.-Z., Ding, T.-B., Wei, D.-D., & Wang, J.-J. (2013). Mining Genes Involved in Insecticide Resistance of Liposcelis bostrychophila Badonnel by Transcriptome and Expression Profile Analysis. PloS one, 8(11). doi: 10.1371/journal.pone.0079878 Douglas, A., Bouvaine, S., & Russell, R. (2011). How the insect immune system interacts with an obligate symbiotic bacterium. Proceedings of the Royal Society B: Biological Sciences, 278(1704), 333-338. Dowen, R. H., Pelizzola, M., Schmitz, R. J., Lister, R., Dowen, J. M., Nery, J. R., . . . Ecker, J. R. (2012). Widespread dynamic DNA methylation in response to biotic stress. Proceedings of the National Academy of Sciences, 109(32), E2183-E2191. 26 Drew, R. E., Settles, M. L., Churchill, E. J., Williams, S. M., Balli, S., & Robison, B. D. (2012). Brain transcriptome variation among behaviorally distinct strains of zebrafish (Danio rerio). BMC genomics, 13. doi: 10.1186/1471-2164-13-323 Dudycha, J. L., Brandon, C. S., & Deitz, K. C. (2012). Population genomics of resource exploitation: insights from gene expression profiles of two Daphnia ecotypes fed alternate resources. Ecology and Evolution, 2(2), 329-340. doi: 10.1002/ece3.30 Dunning, L. T., Dennis, A. B., Park, D., Sinclair, B. J., Newcomb, R. D., & Buckley, T. R. (2013). Identification of cold-responsive genes in a New Zealand alpine stick insect using RNA-Seq. Comparative Biochemistry and Physiology D-Genomics & Proteomics, 8(1), 24-31. doi: 10.1016/j.cbd.2012.10.005 Durban, J., Juarez, P., Angulo, Y., Lomonte, B., Flores-Diaz, M., Alape-Giron, A., . . . Calvete, J. J. (2011). Profiling the venom gland transcriptomes of Costa Rican snakes by 454 pyrosequencing. BMC genomics, 12. doi: 10.1186/1471-2164-12-259 Dworkin, I., Kennerly, E., Tack, D., Hutchinson, J., Brown, J., Mahaffey, J., & Gibson, G. (2009). Genomic Consequences of Background Effects on scalloped Mutant Expressivity in the Wing of Drosophila melanogaster. Genetics, 181(3), 1065-1076. doi: 10.1534/genetics.108.096453 Ekblom, R., Farrell, L. L., Lank, D. B., & Burke, T. (2012). Gene expression divergence and nucleotide differentiation between males of different color morphs and mating strategies in the ruff. Ecology and Evolution, 2(10), 2485-2500. doi: 10.1002/ece3.370 Elmer, K. R., Fan, S., Gunter, H. M., Jones, J. C., Boekhoff, S., Kuraku, S., & Meyer, A. (2010). Rapid evolution and selection inferred from the transcriptomes of sympatric crater lake cichlid fishes. Molecular Ecology, 19, 197-211. doi: 10.1111/j.1365-294X.2009.04488.x Etebari, K., Palfreyman, R. W., Schlipalius, D., Nielsen, L. K., Glatz, R. V., & Asgari, S. (2011). Deep sequencing-based transcriptome analysis of Plutella xylostella larvae parasitized by Diadegma semiclausum. BMC genomics, 12. doi: 10.1186/1471-2164-12-446 Evans, T. G., Chan, F., Menge, B. A., & Hofmann, G. E. (2013). Transcriptomic responses to ocean acidification in larval sea urchins from a naturally variable pH environment. Molecular Ecology, 22(6), 1609-1625. doi: 10.1111/mec.12188 Evans, T. G., Hammill, E., Kaukinen, K., Schulze, A. D., Patterson, D. A., English, K. K., . . . Miller, K. M. (2011). Transcriptomics of environmental acclimatization and survival in wild adult Pacific sockeye salmon (Oncorhynchus nerka) during spawning migration. Molecular Ecology, 20(21), 44724489. doi: 10.1111/j.1365-294X.2011.05276.x Evans, T. G., & Somero, G. N. (2008). A microarray-based transcriptomic time-course of hyper- and hypo-osmotic stress signaling events in the euryhaline fish Gillichthys mirabilis: osmosensors to effectors. Journal of Experimental Biology, 211(22), 3636-3649. doi: 10.1242/jeb.022160 Ewen-Campen, B., Shaner, N., Panfilio, K. A., Suzuki, Y., Roth, S., & Extavour, C. G. (2011). The maternal and early embryonic transcriptome of the milkweed bug Oncopeltus fasciatus. BMC genomics, 12. doi: 10.1186/1471-2164-12-61 Fabro, G., Di Rienzo, J. A., Voigt, C. A., Savchenko, T., Dehesh, K., Somerville, S., & Alvarez, M. E. (2008). Genome-wide expression profiling Arabidopsis at the stage of Golovinomyces cichoracearum haustorium formation. Plant physiology, 146(3), 1421-1439. doi: 10.1104/pp.107.111286 Fan, P., Nie, L., Jiang, P., Feng, J., Lv, S., Chen, X., . . . Li, Y. (2013). Transcriptome Analysis of Salicornia europaea under Saline Conditions Revealed the Adaptive Primary Metabolic Pathways as Early Events to Facilitate Salt Adaptation. PloS one, 8(11). doi: 10.1371/journal.pone.0080595 Fan, S., Elmer, K. R., & Meyer, A. (2012). Genomics of adaptation and speciation in cichlid fishes: recent advances and analyses in African and Neotropical lineages. Royal Society Philosophical Transactions Biological Sciences, 367(1587), 385-394. 27 Fang, G., Passalacqua, K. D., Hocking, J., Llopis, P. M., Gerstein, M., Bergman, N. H., & Jacobs-Wagner, C. (2013). Transcriptomic and phylogenetic analysis of a bacterial cell cycle reveals strong associations between gene co-expression and evolution. BMC genomics, 14. doi: 10.1186/1471-216414-450 Fehsenfeld, S., Kiko, R., Appelhans, Y., Towle, D. W., Zimmer, M., & Melzner, F. (2011). Effects of elevated seawater pCO(2) on gene expression patterns in the gills of the green crab, Carcinus maenas. BMC genomics, 12. doi: 10.1186/1471-2164-12-488 Feldmeyer, B., Elsner, D., & Foitzik, S. (2014). Gene expression patterns associated with caste and reproductive status in ants: worker-specific genes are more derived than queen-specific ones. Molecular Ecology, 23(1), 151-161. doi: 10.1111/mec.12490 Ferreira, P. G., Patalano, S., Chauhan, R., Ffrench-Constant, R., Gabaldon, T., Guigo, R., & Sumner, S. (2013). Transcriptome analyses of primitively eusocial wasps reveal novel insights into the evolution of sociality and the origin of alternative phenotypes. Genome Biology, 14(2), 1-14. Filteau, M., Pavey, S. A., St-Cyr, J., & Bernatchez, L. (2013). Gene Coexpression Networks Reveal Key Drivers of Phenotypic Divergence in Lake Whitefish. Molecular biology and evolution, 30(6), 13841396. doi: 10.1093/molbev/mst053 Fonseca, P., Moreno, R., & Rojo, F. (2011). Growth of Pseudomonas putida at low temperature: global transcriptomic and proteomic analyses. Environmental Microbiology Reports, 3(3), 329-339. doi: 10.1111/j.1758-2229.2010.00229.x Frangipani, E., Perez-Martinez, I., Williams, H. D., Cherbuin, G., & Haas, D. (2014). A novel cyanideinducible gene cluster helps protect Pseudomonas aeruginosa from cyanide. Environmental Microbiology Reports, 6(1), 28-34. doi: 10.1111/1758-2229.12105 Freitak, D., Knorr, E., Vogel, H., & Vilcinskas, A. (2012). Gender-and stressor-specific microRNA expression in Tribolium castaneum. Biology Letters, 8(5), 860-863. Friedrich, M., Chen, R., Daines, B., Bao, R., Caravas, J., Rai, P. K., . . . Peck, S. B. (2011). Phototransduction and clock gene expression in the troglobiont beetle Ptomaphagus hirtus of Mammoth cave. Journal of Experimental Biology, 214(21), 3532-3541. doi: 10.1242/jeb.060368 Fu, B., & He, S. (2012). Transcriptome Analysis of Silver Carp (Hypophthalmichthys molitrix) by PairedEnd RNA Sequencing. DNA Research, 19(2), 131-142. doi: 10.1093/dnares/dsr046 Gallagher, E. P., LaVire, H. M., Bammler, T. K., Stapleton, P. L., Beyer, R. P., & Farin, F. M. (2008). Hepatic expression profiling in smolting and adult coho salmon (Onchorhynchus kisutch). Environmental Research, 106(3), 365-378. doi: 10.1016/j.envres.2007.10.001 Galletti, M. F. B. M., Fujita, A., Nishiyama, M. Y., Jr., Malossi, C. D., Pinter, A., Soares, J. F., . . . Fogaca, A. C. (2013). Natural Blood Feeding and Temperature Shift Modulate the Global Transcriptional Profile of Rickettsia rickettsii Infecting Its Tick Vector. PloS one, 8(10). doi: 10.1371/journal.pone.0077388 Ganot, P., Moya, A., Magnone, V., Allemand, D., Furla, P., & Sabourault, C. (2011). Adaptations to Endosymbiosis in a Cnidarian-Dinoflagellate Association: Differential Gene Expression and Specific Gene Duplications. PLoS genetics, 7(7). doi: 10.1371/journal.pgen.1002187 Garcia-Vedrenne, A. E., Groner, M., Page-Karjian, A., Siegmund, G.-F., Singhal, S., Sziklay, J., & Roberts, S. (2013). Development of Genomic Resources for a thraustochytrid Pathogen and Investigation of Temperature Influences on Gene Expression. PloS one, 8(9), 1-10. Garcia, C., Avila, V., Quesada, H., & Caballero, A. (2012). Gene-expression changes caused by inbreeding protect against inbreeding depression in Drosophila. Genetics, 192(1), 161-172. doi: 10.1534/genetics.112.142687 28 Genissel, A., McIntyre, L. M., Wayne, M. L., & Nuzhdin, S. V. (2008). Cis and trans regulatory effects contribute to natural variation in transcriptome of Drosophila melanogaster. Molecular biology and evolution, 25(1), 101-110. doi: 10.1093/molbev/msm247 Geraldes, A., Pang, J., Thiessen, N., Cezard, T., Moore, R., Zhao, Y., . . . Douglas, C. J. (2011). SNP discovery in black cottonwood (Populus trichocarpa) by population transcriptome resequencing. Mol Ecol Resour, 11, 81-92. doi: 10.1111/j.1755-0998.2010.02960.x Gierga, G., Voss, B., & Hess, W. R. (2012). Non-coding RNAs in marine Synechococcus and their regulation under environmentally relevant stress conditions. The ISME journal, 6(8), 1544-1557. Giger, T., Excoffier, L., Amstutz, U., Day, P. J. R., Champigneulle, A., Hansen, M. M., . . . Largiader, C. R. (2008). Population transcriptomics of life-history variation in the genus Salmo. Molecular Ecology, 17(13), 3095-3108. doi: 10.1111/j.1365-294X.2008.03820.x Gioti, A., Stajich, J. E., & Johannesson, H. (2013). NEUROSPORA AND THE DEAD-END HYPOTHESIS: GENOMIC CONSEQUENCES OF SELFING IN THE MODEL GENUS. Evolution, 67(12), 3600-3616. doi: 10.1111/evo.12206 Gioti, A., Wigby, S., Wertheim, B., Schuster, E., Martinez, P., Pennington, C. J., . . . Chapman, T. (2012). Sex peptide of Drosophila melanogaster males is a global regulator of reproductive processes in females. Proceedings of the Royal Society B-Biological Sciences, 279(1746), 4423-4432. doi: 10.1098/rspb.2012.1634 Goetz, F., Rosauer, D., Sitar, S., Goetz, G., Simchick, C., Roberts, S., . . . Mackenzie, S. (2010). A genetic basis for the phenotypic differentiation between siscowet and lean lake trout (Salvelinus namaycush). Molecular Ecology, 19, 176-196. doi: 10.1111/j.1365-294X.2009.04481.x Goltsev, Y., Rezende, G. L., Vranizan, K., Lanzaro, G., Valle, D., & Levine, M. (2009). Developmental and evolutionary basis for drought tolerance of the Anopheles gambiae embryo. Developmental Biology, 330(2), 462-470. doi: 10.1016/j.ydbio.2009.02.038 Gomulski, L. M., Dimopoulos, G., Xi, Z., Scolari, F., Gabrieli, P., Siciliano, P., . . . Gasperi, G. (2012). Transcriptome Profiling of Sexual Maturation and Mating in the Mediterranean Fruit Fly, Ceratitis capitata. PloS one, 7(1). doi: 10.1371/journal.pone.0030857 Gracey, A. Y., Chaney, M. L., Boomhower, J. P., Tyburczy, W. R., Connor, K., & Somero, G. N. (2008). Rhythms of gene expression in a fluctuating intertidal environment. Current Biology, 18(19), 15011507. Gracey, A. Y., Fraser, E. J., Li, W. Z., Fang, Y. X., Taylor, R. R., Rogers, J., . . . Cossins, A. R. (2004). Coping with cold: An integrative, multitissue analysis of the transcriptome of a poikilothermic vertebrate. Proc Natl Acad Sci U S A, 101(48), 16970-16975. doi: 10.1073/pnas.0403627101 Granados-Cifuentes, C., Bellantuono, A. J., Ridgway, T., Hoegh-Guldberg, O., & Rodriguez-Lanetty, M. (2013). High natural gene expression variation in the reef-building coral Acropora millepora: potential for acclimative and adaptive plasticity. BMC genomics, 14. doi: 10.1186/1471-2164-14-228 Gross, J. B., Furterer, A., Carlson, B. M., & Stahl, B. A. (2013). An Integrated Transcriptome-Wide Analysis of Cave and Surface Dwelling Astyanax mexicanus. PloS one, 8(2). doi: 10.1371/journal.pone.0055659 Gross, S. M., Martin, J. A., Simpson, J., Jazmin Abraham-Juarez, M., Wang, Z., & Visel, A. (2013). De novo transcriptome assembly of drought tolerant CAM plants, Agave deserti and Agave tequilana. BMC genomics, 14. doi: 10.1186/1471-2164-14-563 Grostern, A., & Alvarez-Cohen, L. (2013). RubisCO-based CO2 fixation and C-1 metabolism in the actinobacterium Pseudonocardia dioxanivorans CB1190. Environmental Microbiology, 15(11), 30403053. doi: 10.1111/1462-2920.12144 29 Grumaz, C., Lorenz, S., Stevens, P., Lindemann, E., Schoeck, U., Retey, J., . . . Sohn, K. (2013). Species and condition specific adaptation of the transcriptional landscapes in Candida albicans and Candida dubliniensis. BMC genomics, 14. doi: 10.1186/1471-2164-14-212 Gu, X., Zou, Y., Huang, W., Shen, L., Arendsee, Z., & Su, Z. (2013). Phylogenomic Distance Method for Analyzing Transcriptome Evolution Based on RNA-seq Data. Genome Biology and Evolution, 5(9), 1746-1753. doi: 10.1093/gbe/evt121 Guggisberg, A., Lai, Z., Huang, J., & Rieseberg, L. H. (2013). Transcriptome divergence between introduced and native populations of Canada thistle, Cirsium arvense. New Phytologist, 199(2), 595608. doi: 10.1111/nph.12258 Gui, D., Jia, K., Xia, J., Yang, L., Chen, J., Wu, Y., & Yi, M. (2013). De novo Assembly of the IndoPacific Humpback Dolphin Leucocyte Transcriptome to Identify Putative Genes Involved in the Aquatic Adaptation and Immune Response. PloS one, 8(8). doi: 10.1371/journal.pone.0072417 Gunter, H. M., Fan, S., Xiong, F., Franchini, P., Fruciano, C., & Meyer, A. (2013). Shaping development through mechanical strain: the transcriptional basis of diet-induced phenotypic plasticity in a cichlid fish. Molecular Ecology, 22(17), 4516-4531. doi: 10.1111/mec.12417 Guo, B., Chain, F. J. J., Bornberg-Bauer, E., Leder, E. H., & Merila, J. (2013). Genomic divergence between nine- and three-spined sticklebacks. BMC genomics, 14, 1-11. Hall, D. E., Yuen, M. M. S., Jancsik, S., Quesada, A. L., Dullat, H. K., Li, M., . . . Bohlmann, J. (2013). Transcriptome resources and functional characterization of monoterpene synthases for two host species of the mountain pine beetle, lodgepole pine (Pinus contorta) and jack pine (Pinus banksiana). Bmc Plant Biology, 13. doi: 10.1186/1471-2229-13-80 Hanson, S. J., Stelzer, C.-P., Welch, D. B. M., & Logsdon, J. M., Jr. (2013). Comparative transcriptome analysis of obligately asexual and cyclically sexual rotifers reveals genes with putative functions in sexual reproduction, dormancy, and asexual egg production. BMC genomics, 14, 1-17. Harms, L., Frickenhaus, S., Schiffer, M., Mark, F. C., Storch, D., Poertner, H.-O., . . . Lucassen, M. (2013). Characterization and analysis of a transcriptome from the boreal spider crab Hyas araneus. Comparative Biochemistry and Physiology D-Genomics & Proteomics, 8(4), 344-351. doi: 10.1016/j.cbd.2013.09.004 Harr, B., & Turner, L. M. (2010). Genome-wide analysis of alternative splicing evolution among Mus subspecies. Molecular Ecology, 19, 228-239. doi: 10.1111/j.1365-294X.2009.04490.x Harris, S. E., Munshi-South, J., Obergfell, C., & O'Neill, R. (2013). Signatures of Rapid Evolution in Urban and Rural Transcriptomes of White-Footed Mice (Peromyscus leucopus) in the New York Metropolitan Area. PloS one, 8(8). doi: 10.1371/journal.pone.0074938 He, Z., Zhou, A., Baidoo, E., He, Q., Joachimiak, M. P., Benke, P., . . . Zhou, J. (2010). Global Transcriptional, Physiological, and Metabolite Analyses of the Responses of Desulfovibrio vulgaris Hildenborough to Salt Adaptation. Applied and Environmental Microbiology, 76(5), 1574-1586. doi: 10.1128/aem.02141-09 Healy, T. M., Tymchuk, W. E., Osborne, E. J., & Schulte, P. M. (2010). Heat shock response of killifish (Fundulus heteroclitus): candidate gene and heterologous microarray approaches. Physiological genomics, 41(2), 171-184. Heath, K. D., Burke, P. V., & Stinchcombe, J. R. (2012). Coevolutionary genetic variation in the legumerhizobium transcriptome. Molecular Ecology, 21(19), 4735-4747. doi: 10.1111/j.1365294X.2012.05629.x Hegarty, M. J., Barker, G. L., Brennan, A. C., Edwards, K. J., Abbott, R. J., & Hiscock, S. J. (2009). Extreme changes to gene expression associated with homoploid hybrid speciation. Molecular Ecology, 18(5), 877-889. doi: 10.1111/j.1365-294X.2008.04054.x 30 Hegarty, M. J., Jones, J. M., Wilson, I. D., Barker, G. L., Coghill, J. A., Sanchez-Baracaldo, P., . . . Hiscock, S. J. (2005). Development of anonymous cDNA microarrays to study changes to the Senecio floral transcriptome during hybrid speciation. Molecular Ecology, 14(8), 2493-2510. doi: 10.1111/j.1365-294X.2005.02608.x Henning, F., Jones, J. C., Franchini, P., & Meyer, A. (2013). Transcriptomics of morphological color change in polychromatic Midas cichlids. BMC genomics, 14. doi: 10.1186/1471-2164-14-171 Herranz, R., Larkin, O. J., Hill, R. J. A., Lopez-Vidriero, I., van Loon, J. J. W. A., & Javier Medina, F. (2013). Suboptimal evolutionary novel environments promote singular altered gravity responses of transcriptome during Drosophila metamorphosis. Bmc Evolutionary Biology, 13. doi: 10.1186/14712148-13-133 Hershkovitz, V., Sela, N., Taha-Salaime, L., Liu, J., Rafael, G., Kessler, C., . . . Droby, S. (2013). Denovo assembly and characterization of the transcriptome of Metschnikowia fructicola reveals differences in gene expression following interaction with Penicillium digitatum and grapefruit peel. BMC genomics, 14. doi: 10.1186/1471-2164-14-168 Hewson, I., Poretsky, R. S., Beinart, R. A., White, A. E., Shi, T., Bench, S. R., . . . Zehr, J. P. (2009). In situ transcriptomic analysis of the globally important keystone N-2-fixing taxon Crocosphaera watsonii. Isme Journal, 3(5), 618-631. doi: 10.1038/ismej.2009.8 Hines, H. M., Papa, R., Ruiz, M., Papanicolaou, A., Wang, C., Nijhout, H. F., . . . Reed, R. D. (2012). Transcriptome analysis reveals novel patterning and pigmentation genes underlying Heliconius butterfly wing pattern variation. BMC genomics, 13. doi: 10.1186/1471-2164-13-288 Hocher, V., Alloisio, N., Auguy, F., Fournier, P., Doumas, P., Pujic, P., . . . Bogusz, D. (2011). Transcriptomics of Actinorhizal Symbioses Reveals Homologs of the Whole Common Symbiotic Signaling Cascade. Plant physiology, 156(2), 700-711. doi: 10.1104/pp.111.174151 Hodgins, K. A., Lai, Z., Oliveira, L. O., Still, D. W., Scascitelli, M., Barker, M. S., . . . Rieseberg, L. H. (2014). Genomics of Compositae crops: reference transcriptome assemblies and evidence of hybridization with wild relatives. Mol Ecol Resour, 14(1), 166-177. doi: 10.1111/1755-0998.12163 Hoffman, J. I. (2011). Gene discovery in the Antarctic fur seal (Arctocephalus gazella) skin transcriptome. Mol Ecol Resour, 11(4), 703-710. doi: 10.1111/j.1755-0998.2011.02999.x Hohnjec, N., Vieweg, M. E., Puhler, A., Becker, A., & Kuster, H. (2005). Overlaps in the transcriptional profiles of Medicago truncatula roots inoculated with two different Glomus fungi provide insights into the genetic program activated during arbuscular mycorrhiza. Plant physiology, 137(4), 1283-1301. doi: 10.1104/pp.104.056572 Holliday, J. A., Ralph, S. G., White, R., Bohlmann, J., & Aitken, S. N. (2008). Global monitoring of autumn gene expression within and among phenotypically divergent populations of Sitka spruce (Picea sitchensis). New Phytologist, 178(1), 103-122. doi: 10.1111/j.1469-8137.2007.02346.x Hori, T. S., Gamperl, A. K., Nash, G., Booman, M., Barat, A., & Rise, M. L. (2013). The impact of a moderate chronic temperature increase on spleen immune-relevant gene transcription depends on whether Atlantic cod (Gadus morhua) are stimulated with bacterial versus viral antigens. Genome, 56(10), 567-576. doi: 10.1139/gen-2013-0090 Horvath, D. P., Soto-Suarez, M., Chao, W. S., Jia, Y., & Anderson, J. V. (2005). Transcriptome analysis of paradormancy release in root buds of leafy spurge (Euphorbia esula). Weed Science, 53(6), 795801. doi: 10.1614/ws-05-066r1.1 Hou, Z.-C., Sterner, K. N., Romero, R., Than, N. G., Gonzalez, J. M., Weckle, A., . . . Wildman, D. E. (2012). Elephant Transcriptome Provides Insights into the Evolution of Eutherian Placentation. Genome Biology and Evolution, 4(5), 713-725. doi: 10.1093/gbe/evs045 Hoy, M. A., Yu, F., Meyer, J. M., Tarazona, O. A., Jeyaprakash, A., & Wu, K. (2013). Transcriptome sequencing and annotation of the predatory mite Metaseiulus occidentalis (Acari: Phytoseiidae): a 31 cautionary tale about possible contamination by prey sequences. Experimental and Applied Acarology, 59(3), 283-296. doi: 10.1007/s10493-012-9603-4 Hull, J. J., Geib, S. M., Fabrick, J. A., & Brent, C. S. (2013). Sequencing and De Novo Assembly of the Western Tarnished Plant Bug (Lygus hesperus) Transcriptome. PloS one, 8(1). doi: 10.1371/journal.pone.0055105 Hüning, A. K., Melzner, F., Thomsen, J., Gutowska, M. A., Krämer, L., Frickenhaus, S., . . . Lucassen, M. (2013). Impacts of seawater acidification on mantle gene expression patterns of the Baltic Sea blue mussel: implications for shell formation and energy metabolism. Marine biology, 160(8), 1845-1861. Huth, T. J., & Place, S. P. (2013). De novo assembly and characterization of tissue specific transcriptomes in the emerald notothen, Trematomus bernacchii. BMC genomics, 14, 1-13. Hwang, Y.-S., Jung, G., & Jin, E. (2008). Transcriptome analysis of acclimatory responses to thermal stress in Antarctic algae. Biochemical and Biophysical Research Communications, 367(3), 635-641. doi: 10.1016/j.bbrc.2007.12.176 Ibarra-Laclette, E., Albert, V. A., Perez-Torres, C. A., Zamudio-Hernandez, F., Ortega-Estrada, M. d. J., Herrera-Estrella, A., & Herrera-Estrella, L. (2011). Transcriptomics and molecular evolutionary rate analysis of the bladderwort (Utricularia), a carnivorous plant with a minimal genome. Bmc Plant Biology, 11. doi: 10.1186/1471-2229-11-101 Ignatov, D. V., Skvortsov, T. A., Majorov, K. B., Apt, A. S., & Azhikina, T. L. (2010). Adaptive Changes in Mycobacterium avium Gene Expression Profile Following Infection of Genetically Susceptible and Resistant Mice. Acta Naturae, 2(3), 78-84. Immonen, E., & Ritchie, M. G. (2012). The genomic response to courtship song stimulation in female Drosophila melanogaster. Proceedings of the Royal Society B: Biological Sciences, 279(1732), 13591365. Isanapong, J., Hambright, W. S., Willis, A. G., Boonmee, A., Callister, S. J., Burnum, K. E., . . . Rodrigues, J. L. M. (2013). Development of an ecophysiological model for Diplosphaera colotermitum TAV2, a termite hindgut Verrucomicrobium. Isme Journal, 7(9), 1803-1813. doi: 10.1038/ismej.2013.74 Jammes, F., Lecomte, P., Almeida-Engler, J., Bitton, F., Martin-Magniette, M. L., Renou, J. P., . . . Favery, B. (2005). Genome-wide expression profiling of the host response to root-knot nematode infection in Arabidopsis. Plant Journal, 44(3), 447-458. doi: 10.1111/j.1365-313X.2005.02532.x Jansen, M., Vergauwen, L., Vandenbrouck, T., Knapen, D., Dom, N., Spanier, K. I., . . . De Meester, L. (2013). Gene expression profiling of three different stressors in the water flea Daphnia magna. Ecotoxicology, 22(5), 900-914. Janz, D., Behnke, K., Schnitzler, J.-P., Kanawati, B., Schmitt-Kopplin, P., & Polle, A. (2010). Pathway analysis of the transcriptome and metabolome of salt sensitive and tolerant poplar species reveals evolutionary adaption of stress tolerance mechanisms. Bmc Plant Biology, 10. doi: 10.1186/14712229-10-150 Jeffries, K. M., Hinch, S. G., Sierocinski, T., Clark, T. D., Eliason, E. J., Donaldson, M. R., . . . Miller, K. M. (2012). Consequences of high temperatures and premature mortality on the transcriptome and blood physiology of wild adult sockeye salmon (Oncorhynchus nerka). Ecology and Evolution, 2(7), 1747-1764. doi: 10.1002/ece3.274 Jeffries, K. M., Hinch, S. G., Sierocinski, T., Pavlidis, P., & Miller, K. M. (2014). Transcriptomic responses to high water temperature in two species of Pacific salmon. Evolutionary Applications, 7(2), 286-300. doi: 10.1111/eva.12119 Jeukens, J., Renaut, S., St-Cyr, J., Nolte, A. W., & Bernatchez, L. (2010). The transcriptomics of sympatric dwarf and normal lake whitefish (Coregonus clupeaformis spp., Salmonidae) divergence as revealed by next-generation sequencing. Molecular Ecology, 19(24), 5389-5403. doi: 10.1111/j.1365294X.2010.04934.x 32 Jeyasingh, P. D., Ragavendran, A., Paland, S., Lopez, J. A., Sterner, R. W., & Colbourne, J. K. (2011). How do consumers deal with stoichiometric constraints? Lessons from functional genomics using Daphnia pulex. Molecular Ecology, 20(11), 2341-2352. doi: 10.1111/j.1365-294X.2011.05102.x Jiang, Z.-F., & Machado, C. A. (2009). Evolution of Sex-Dependent Gene Expression in Three Recently Diverged Species of Drosophila. Genetics, 183(3), 1175-1185. doi: 10.1534/genetics.109.105775 Jimenez-Guri, E., Huerta-Cepas, J., Cozzuto, L., Wotton, K. R., Kang, H., Himmelbauer, H., . . . Jaeger, J. (2013). Comparative transcriptomics of early dipteran development. BMC genomics, 14, 1-17. Johansson, F., Veldhoen, N., Lind, M. I., & Helbing, C. C. (2013). Phenotypic plasticity in the hepatic transcriptome of the European common frog (Rana temporaria): the interplay between environmental induction and geographical lineage on developmental response. Molecular Ecology, 22(22), 56085623. doi: 10.1111/mec.12497 Johansson, H., Dhaygude, K., Lindstrom, S., Helantera, H., Sundstrom, L., & Trontti, K. (2013). A Metatranscriptomic Approach to the Identification of Microbiota Associated with the Ant Formica exsecta. PloS one, 8(11). doi: 10.1371/journal.pone.0079777 Johnson, J. G., Morey, J. S., Neely, M. G., Ryan, J. C., & Van Dolah, F. M. (2012). Transcriptome remodeling associated with chronological aging in the dinoflagellate, Karenia brevis. Mar Genomics, 5, 15-25. doi: 10.1016/j.margen.2011.08.005 Jouaux, A., Lafont, M., Blin, J.-L., Houssin, M., Mathieu, M., & Lelong, C. (2013). Physiological change under OsHV-1 contamination in Pacific oyster Crassostrea gigas through massive mortality events on fields. BMC genomics, 14, 15-28. Kaakoush, N. O., Baar, C., MacKichan, J., Schmidt, P., Fox, E. M., Schuster, S. C., & Mendz, G. L. (2009). Insights into the molecular basis of the microaerophily of three Campylobacterales: a comparative study. Antonie Van Leeuwenhoek International Journal of General and Molecular Microbiology, 96(4), 545-557. doi: 10.1007/s10482-009-9370-3 Kadam, S. V., Wegener-Feldbruegge, S., Sogaard-Andersen, L., & Velicer, G. J. (2008). Novel transcriptome patterns accompany evolutionary restoration of defective social development in the bacterium Myxococcus xanthus. Molecular biology and evolution, 25(7), 1274-1281. doi: 10.1093/molbev/msn076 Kalyuzhnaya, M. G., Beck, D. A. C., Suciu, D., Pozhitkov, A., Lidstrom, M. E., & Chistoserdova, L. (2010). Functioning in situ: gene expression in Methylotenera mobilis in its native environment as assessed through transcriptomics. Isme Journal, 4(3), 388-398. doi: 10.1038/ismej.2009.117 Kanter, U., Heller, W., Durner, J., Winkler, J. B., Engel, M., Behrendt, H., . . . Ernst, D. (2013). Molecular and Immunological Characterization of Ragweed (Ambrosia artemisiifolia L.) Pollen after Exposure of the Plants to Elevated Ozone over a Whole Growing Season. PloS one, 8(4). doi: 10.1371/journal.pone.0061518 Kao, D., Felix, D., & Aboobaker, A. (2013). The planarian regeneration transcriptome reveals a shared but temporally shifted regulatory program between opposing head and tail scenarios. BMC genomics, 14. doi: 10.1186/1471-2164-14-797 Kao, K. C., & Sherlock, G. (2008). Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nature genetics, 40(12), 1499-1504. doi: 10.1038/ng.280 Kaplinsky, N. J., Gilbert, S. F., Cebra-Thomas, J., Lillevaeli, K., Saare, M., Chang, E. Y., . . . Watson, R. (2013). The Embryonic Transcriptome of the Red-Eared Slider Turtle (Trachemys scripta). PloS one, 8(6). doi: 10.1371/journal.pone.0066357 Kariuki, M. M., Hearne, L. B., & Beerntsen, B. T. (2010). Differential transcript expression between the microfilariae of the filarial nematodes, Brugia malayi and B. pahangi. BMC genomics, 11. doi: 10.1186/1471-2164-11-225 33 Kassahn, K. S., Caley, M. J., Ward, A. C., Connolly, A. R., Stone, G., & Crozier, R. H. (2007). Heterologous microarray experiments used to identify the early gene response to heat stress in a coral reef fish. Molecular Ecology, 16(8), 1749-1763. doi: 10.1111/j.1365-294X.2006.03178.x Katoh, H., Asthana, R., & Ohmori, M. (2004). Gene expression in the cyanobacterium Anabaena sp. PCC7120 under desiccation. Microbial Ecology, 47(2), 164-174. Kawanabe, T., Fujimoto, R., Sasaki, T., Taylor, J. M., & Dennis, E. S. (2012). A comparison of transcriptome and epigenetic status between closely related species in the genus Arabidopsis. Gene, 506(2), 301-309. doi: 10.1016/j.gene.2012.07.003 Keeling, C. I., Henderson, H., Li, M., Yuen, M., Clark, E. L., Fraser, J. D., . . . Bohlmann, J. (2012). Transcriptome and full-length cDNA resources for the mountain pine beetle, Dendroctonus ponderosae Hopkins, a major insect pest of pine forests. Insect Biochemistry and Molecular Biology, 42(8), 525-536. doi: 10.1016/j.ibmb.2012.03.010 Kegel, J. U., John, U., Valentin, K., & Frickenhaus, S. (2013). Genome Variations Associated with Viral Susceptibility and Calcification in Emiliania huxleyi. PloS one, 8(11). doi: 10.1371/journal.pone.0080684 Kelley, J. L., Passow, C. N., Plath, M., Arias Rodriguez, L., Yee, M.-C., & Tobler, M. (2012). Genomic resources for a model in adaptation and speciation research: characterization of the Poecilia mexicana transcriptome. BMC genomics, 13, 1-13. Kempema, L. A., Cui, X., Holzer, F. M., & Walling, L. L. (2007). Arabidopsis transcriptome changes in response to phloem-feeding silverleaf whitefly nymphs. Similarities and distinctions in responses to aphids. Plant physiology, 143(2), 849-865. doi: 10.1104/pp.106.090662 Kersten, B., Ghirardo, A., Schnitzler, J.-P., Kanawati, B., Schmitt-Kopplin, P., Fladung, M., & Schroeder, H. (2013). Integrated transcriptomics and metabolomics decipher differences in the resistance of pedunculate oak to the herbivore Tortrix viridana L. BMC genomics, 14. doi: 10.1186/1471-2164-14737 Khaitovich, P., Hellmann, I., Enard, W., Nowick, K., Leinweber, M., Franz, H., . . . Paabo, S. (2005). Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science, 309(5742), 1850-1854. doi: 10.1126/science.1108296 Kijimoto, T., Costello, J., Tang, Z., Moczek, A. P., & Andrews, J. (2009). EST and microarray analysis of horn development in Onthophagus beetles. BMC genomics, 10. doi: 10.1186/1471-2164-10-504 Kim, J., Carlos Oliveros, J., Nikel, P. I., de Lorenzo, V., & Silva-Rocha, R. (2013). Transcriptomic fingerprinting of Pseudomonas putida under alternative physiological regimes. Environmental Microbiology Reports, 5(6), 883-891. doi: 10.1111/1758-2229.12090 Kim, M., Park, S., Polle, J. E. W., & Jin, E. (2010). Gene expression profiling of Dunaliella sp acclimated to different salinities. Phycological Research, 58(1), 17-28. doi: 10.1111/j.1440-1835.2009.00554.x Kimura, H., Young, C. R., Martinez, A., & DeLong, E. F. (2011). Light-induced transcriptional responses associated with proteorhodopsin-enhanced growth in a marine flavobacterium. Isme Journal, 5(10), 1641-1651. doi: 10.1038/ismej.2011.36 Kitano, J., Kawagishi, Y., Mori, S., Peichel, C. L., Makino, T., Kawata, M., & Kusakabe, M. (2011). Divergence in Sex Steroid Hormone Signaling between Sympatric Species of Japanese Threespine Stickleback. PloS one, 6(12). doi: 10.1371/journal.pone.0029253 Kobayashi, M. J., Takeuchi, Y., Kenta, T., Kume, T., Diway, B., & Shimizu, K. K. (2013). Mass flowering of the tropical tree Shorea beccariana was preceded by expression changes in flowering and drought-responsive genes. Molecular Ecology, 22(18), 4767-4782. doi: 10.1111/mec.12344 Konotchick, T., Dupont, C. L., Valas, R. E., Badger, J. H., & Allen, A. E. (2013). Transcriptomic analysis of metabolic function in the giant kelp, Macrocystis pyrifera, across depth and season. New Phytologist, 198(2), 398-407. doi: 10.1111/nph.12160 34 Kozak, G. M., Brennan, R. S., Berdan, E. L., Fuller, R. C., & Whitehead, A. (2014). Functional and population genomic divergence within and between two species of killifish adapted to different osmotic niches. Evolution, 68(1), 63-80. doi: 10.1111/evo.12265 Künstner, A., Wolf, J. B., Backström, N., Whitney, O., Balakrishnan, C. N., Day, L., ... & Ellegren, H. (2010). Comparative genomics based on massive parallel transcriptome sequencing reveals patterns of substitution and selection across 10 bird species. Molecular Ecology, 19(s1), 266-276.Kusnierczyk, A., Winge, P., Jorstad, T. S., Troczynska, J., Rossiter, J. T., & Bones, A. M. (2008). Towards global understanding of plant defence against aphids - timing and dynamics of early Arabidopsis defence responses to cabbage aphid (Brevicoryne brassicae) attack. Plant Cell and Environment, 31(8), 10971115. doi: 10.1111/j.1365-3040.2008.01823.x Kuśnierczyk, A., Winge, P. E. R., JØRSTAD, T. S., TROCZYŃSKA, J., Rossiter, J. T., & Bones, A. M. (2008). Towards global understanding of plant defence against aphids–timing and dynamics of early Arabidopsis defence responses to cabbage aphid (Brevicoryne brassicae) attack. Plant, cell & environment, 31(8), 1097-1115. Küster, H., Becker, A., Firnhaber, C., Hohnjec, N., Manthey, K., Perlick, A. M., . . . Goesmann, A. (2007). Development of bioinformatic tools to support EST-sequencing,< i> in silico</i>-and microarraybased transcriptome profiling in mycorrhizal symbioses. Phytochemistry, 68(1), 19-32. Kvist, J., Wheat, C. W., Kallioniemi, E., Saastamoinen, M., Hanski, I., & Frilander, M. J. (2013). Temperature treatments during larval development reveal extensive heritable and plastic variation in gene expression and life history traits. Molecular Ecology, 22(3), 602-619. Lai, Z., Kane, N. C., Zou, Y., & Rieseberg, L. H. (2008). Natural variation in gene expression between wild and weedy populations of Helianthus annuus. Genetics, 179(4), 1881-1890. Lang, R. P., Bayne, C. J., Camara, M. D., Cunningham, C., Jenny, M. J., & Langdon, C. J. (2009). Transcriptome Profiling of Selectively Bred Pacific Oyster Crassostrea gigas Families that Differ in Tolerance of Heat Shock. Marine Biotechnology, 11(5), 650-668. doi: 10.1007/s10126-009-9181-6 Latta, L. C., Weider, L. J., Colbourne, J. K., & Pfrender, M. E. (2012). The evolution of salinity tolerance in Daphnia: a functional genomics approach. Ecology Letters, 15(8), 794-802. Le Trionnaire, G., Francis, F., Jaubert-Possamai, S., Bonhomme, J., De Pauw, E., Gauthier, J. P., . . . Tagu, D. (2009). Transcriptomic and proteomic analyses of seasonal photoperiodism in the pea aphid. BMC genomics, 10(456), 1-14. Le Trionnaire, G., Jaubert-Possamai, S., Bonhomme, J., Gauthier, J.-P., Guernec, G., Le Cam, A., . . . Tagu, D. (2012). Transcriptomic profiling of the reproductive mode switch in the pea aphid in response to natural autumnal photoperiod. Journal of Insect Physiology, 58(12), 1517-1524. doi: 10.1016/j.jinsphys.2012.07.009 Leakey, A. D. B., Ainsworth, E. A., Bernard, S. M., Markelz, R. J. C., Ort, D. R., Placella, S. A., . . . Yuan, S. (2009). Gene expression profiling: opening the black box of plant ecosystem responses to global change. Global Change Biology, 15(5), 1201-1213. doi: 10.1111/j.1365-2486.2008.01818.x Lee, J. H., Choi, J. Y., Tao, X. Y., Kim, J. S., Kim, W., & Je, Y. H. (2013). Transcriptome Analysis of the Small Brown Planthopper, Laodelphax striatellus Carrying Rice stripe virus. Plant Pathology Journal, 29(3), 330-337. doi: 10.5423/ppj.nt.01.2013.0001 Lee, S. F., Chen, Z., McGrath, A., Good, R. T., & Batterham, P. (2011). Identification, analysis, and linkage mapping of expressed sequence tags from the Australian sheep blowfly. BMC genomics, 12. doi: 10.1186/1471-2164-12-406 Lehnert, E. M., Burriesci, M. S., & Pringle, J. R. (2012). Developing the anemone Aiptasia as a tractable model for cnidarian-dinoflagellate symbiosis: the transcriptome of aposymbiotic A. pallida. BMC genomics, 13. doi: 10.1186/1471-2164-13-271 35 Lei, Y., Zhu, X., Xie, W., Wu, Q., Wang, S., Guo, Z., . . . Zhang, Y. (2014). Midgut transcriptome response to a Cry toxin in the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). Gene, 533(1), 180-187. doi: 10.1016/j.gene.2013.09.091 Lemay, M. A., Donnelly, D. J., & Russello, M. A. (2013). Transcriptome-wide comparison of sequence variation in divergent ecotypes of kokanee salmon. BMC genomics, 14. doi: 10.1186/1471-2164-14308 Lemay, M. A., Henry, P., Lamb, C. T., Robson, K. M., & Russello, M. A. (2013). Novel genomic resources for a climate change sensitive mammal: characterization of the American pika transcriptome. BMC genomics, 14. doi: 10.1186/1471-2164-14-311 Lemmetyinen, J., Piironen, J., Kiiskinen, P., Hassinen, M., & Vornanen, M. (2013). Comparison of gene expression in the gill of salmon (Salmo salar) smolts from anadromous and landlocked populations. Paper presented at the Annales Zoologici Fennici. Lewandowska-Sabat, A. M., Winge, P., Fjellheim, S., Dørum, G., Bones, A. M., & Rognli, O. A. (2012). Genome wide transcriptional profiling of acclimation to photoperiod in high-latitude accessions of< i> Arabidopsis thaliana</i>. Plant Science, 185, 143-155. Li, A., Fang, M. D., Song, W. Q., Chen, C. B., Qi, L. W., & Wang, C. G. (2012). Gene expression profiles of two intraspecific Larix lines and their reciprocal hybrids. Mol Biol Rep, 39(4), 3773-3784. doi: 10.1007/s11033-011-1154-y Li, B.-W., Wang, Z., Rush, A. C., Mitreva, M., & Weil, G. J. (2012). Transcription profiling reveals stageand function-dependent expression patterns in the filarial nematode Brugia malayi. BMC genomics, 13(1), 184. Li, J., Li, X., Chen, Y., Yang, Z., & Guo, S. (2012). Solexa sequencing based transcriptome analysis of Helicoverpa armigera larvae. Molecular Biology Reports, 39(12), 11051-11059. doi: 10.1007/s11033012-2008-y Li, S.-W., Yang, H., Liu, Y.-F., Liao, Q.-R., Du, J., & Jin, D.-C. (2012). Transcriptome and Gene Expression Analysis of the Rice Leaf Folder, Cnaphalocrosis medinalis. PloS one, 7(11). doi: 10.1371/journal.pone.0047401 Li, T., & Brouwer, M. (2013). Field study of cyclic hypoxic effects on gene expression in grass shrimp hepatopancreas. Comparative Biochemistry and Physiology D-Genomics & Proteomics, 8(4), 309316. doi: 10.1016/j.cbd.2013.09.001 Li, Z.-Q., Zhang, S., Ma, Y., Luo, J.-Y., Wang, C.-Y., Lv, L.-M., . . . Cui, J.-J. (2013). First Transcriptome and Digital Gene Expression Analysis in Neuroptera with an Emphasis on Chemoreception Genes in Chrysopa pallens (Rambur). PloS one, 8(6). doi: 10.1371/journal.pone.0067151 Lin, H.-C., Wong, Y. H., Tsang, L. M., Chu, K. H., Qian, P.-Y., & Chan, B. K. K. (2014). First study on gene expression of cement proteins and potential adhesion-related genes of a membranous-based barnacle as revealed from Next-Generation Sequencing technology. Biofouling, 30(2), 169-181. doi: 10.1080/08927014.2013.853051 Liu, G., Mattick, J. S., & Taft, R. J. (2013). A meta-analysis of the genomic and transcriptomic composition of complex life. Cell Cycle, 12(13), 2061-2072. doi: 10.4161/cc.25134 Liu, S., Chougule, N. P., Vijayendran, D., & Bonning, B. C. (2012). Deep Sequencing of the Transcriptomes of Soybean Aphid and Associated Endosymbionts. PloS one, 7(9). doi: 10.1371/journal.pone.0045161 Liu, Z., Ma, L., Nan, Z., & Wang, Y. (2013). Comparative Transcriptional Profiling Provides Insights into the Evolution and Development of the Zygomorphic Flower of Vicia sativa (Papilionoideae). PloS one, 8(2). doi: 10.1371/journal.pone.0057338 36 Lockwood, B. L., Sanders, J. G., & Somero, G. N. (2010). Transcriptomic responses to heat stress in invasive and native blue mussels (genus Mytilus): molecular correlates of invasive success. Journal of Experimental Biology, 213(20), 3548-3558. doi: 10.1242/jeb.046094 Lockwood, B. L., & Somero, G. N. (2011). Transcriptomic responses to salinity stress in invasive and native blue mussels (genus Mytilus). Molecular Ecology, 20(3), 517-529. doi: 10.1111/j.1365294X.2010.04973.x Logacheva, M. D., Kasianov, A. S., Vinogradov, D. V., Samigullin, T. H., Gelfand, M. S., Makeev, V. J., & Penin, A. A. (2011). De novo sequencing and characterization of floral transcriptome in two species of buckwheat (Fagopyrum). BMC genomics, 12. doi: 10.1186/1471-2164-12-30 Lorenz, P., Dietmann, S., Wilhelm, T., Koczan, D., Autran, S., Gad, S., . . . Thiesen, H.-J. (2010). The ancient mammalian KRAB zinc finger gene cluster on human chromosome 8q24.3 illustrates principles of C2H2 zinc finger evolution associated with unique expression profiles in human tissues. BMC genomics, 11. doi: 10.1186/1471-2164-11-206 Lowe, C. D., Mello, L. V., Samatar, N., Martin, L. E., Montagnes, D. J. S., & Watts, P. C. (2011). The transcriptome of the novel dinoflagellate Oxyrrhis marina (Alveolata: Dinophyceae): response to salinity examined by 454 sequencing. BMC genomics, 12. doi: 10.1186/1471-2164-12-519 Ludeman, D. A., Farrar, N., Riesgo, A., Paps, J., & Leys, S. P. (2014). Evolutionary origins of sensation in metazoans: functional evidence for a new sensory organ in sponges. Bmc Evolutionary Biology, 14. doi: 10.1186/1471-2148-14-3 Lundberg, M., Boss, J., Canback, B., Liedvogel, M., Larson, K. W., Grahn, M., . . . Wright, A. (2013). Characterisation of a transcriptome to find sequence differences between two differentially migrating subspecies of the willow warbler Phylloscopus trochilus. BMC genomics, 14. doi: 10.1186/14712164-14-330 Lv, J., Liu, P., Wang, Y., Gao, B., Chen, P., & Li, J. (2013). Transcriptome Analysis of Portunus trituberculatus in Response to Salinity Stress Provides Insights into the Molecular Basis of Osmoregulation. PloS one, 8(12). doi: 10.1371/journal.pone.0082155 Maaty, W. S., Wiedenheft, B., Tarlykov, P., Schaff, N., Heinemann, J., Robison-Cox, J., . . . Bothner, B. (2009). Something Old, Something New, Something Borrowed; How the Thermoacidophilic Archaeon Sulfolobus solfataricus Responds to Oxidative Stress. PloS one, 4(9). doi: 10.1371/journal.pone.0006964 Malenke, J. R., Milash, B., Miller, A. W., & Dearing, M. D. (2013). Transcriptome sequencing and microarray development for the woodrat (Neotoma spp.): custom genetic tools for exploring herbivore ecology. Mol Ecol Resour, 13(4), 674-687. doi: 10.1111/1755-0998.12092 Malone, J. H., Hawkins, D. L., Jr., & Michalak, P. (2006). Sex-biased gene expression in a ZW sex determination system. Journal of Molecular Evolution, 63(4), 427-436. doi: 10.1007/s00239-0050263-4 Mancia, A., Ryan, J. C., Chapman, R. W., Wu, Q., Warr, G. W., Gulland, F., & Van Dolah, F. M. (2012). Health status, infection and disease in California sea lions (< i> Zalophus californianus</i>) studied using a canine microarray platform and machine-learning approaches. Developmental & Comparative Immunology, 36(4), 629-637. Mancia, A., Warr, G. W., Almeida, J. S., Veloso, A., Wells, R. S., & Chapman, R. W. (2010). Transcriptome Profiles: Diagnostic Signature of Dolphin Populations. Estuaries and Coasts, 33(4), 919-929. doi: 10.1007/s12237-010-9287-0 Mancia, A., Warr, W., & Chapman, R. W. (2008). A transcriptomic analysis of the stress induced by capture-release health assessment studies in wild dolphins (Tursiops truncatus). Molecular Ecology, 17(11), 2581-2589. doi: 10.1111/j.1365-294X.2008.03784.x 37 Mane, S. P., Vasquez-Robinet, C., Sioson, A. A., Heath, L. S., & Grene, R. (2007). Early PLD alphamediated events in response to progressive drought stress in Arabidopsis: a transcriptome analysis. Journal of Experimental Botany, 58(2), 241-252. doi: 10.1093/jxb/erl262 Manfredini, F., Lucas, C., Nicolas, M., Keller, L., Shoemaker, D., & Grozinger, C. M. (2014). Molecular and social regulation of worker division of labour in fire ants. Molecular Ecology, 23(3), 660-672. doi: 10.1111/mec.12626 Manousaki, T., Hull, P. M., Kusche, H., Machado-Schiaffino, G., Franchini, P., Harrod, C., . . . Meyer, A. (2013). Parsing parallel evolution: ecological divergence and differential gene expression in the adaptive radiations of thick-lipped Midas cichlid fishes from Nicaragua. Molecular Ecology, 22(3), 650-669. doi: 10.1111/mec.12034 Margres, M. J., Aronow, K., Loyacano, J., & Rokyta, D. R. (2013). The venom-gland transcriptome of the eastern coral snake (Micrurus fulvius) reveals high venom complexity in the intragenomic evolution of venoms. BMC genomics, 14, 1-18. Marinkovic, M., de Leeuw, W. C., de Jong, M., Kraak, M. H. S., Admiraal, W., Breit, T. M., & Jonker, M. J. (2012). Combining Next-Generation Sequencing and Microarray Technology into a Transcriptomics Approach for the Non-Model Organism Chironomus riparius. PloS one, 7(10). doi: 10.1371/journal.pone.0048096 Marques, I. J., Leito, J. T. D., Spaink, H. P., Testerink, J., Jaspers, R. T., Witte, F., . . . Bagowski, C. P. (2008). Transcriptome analysis of the response to chronic constant hypoxia in zebrafish hearts. Journal of Comparative Physiology B-Biochemical Systemic and Environmental Physiology, 178(1), 77-92. doi: 10.1007/s00360-007-0201-4 Martens-Uzunova, E. S., & Schaap, P. J. (2008). An evolutionary conserved D-galacturonic acid metabolic pathway operates across filamentous fungi capable of pectin degradation. Fungal Genetics and Biology, 45(11), 1449-1457. doi: 10.1016/j.fgb.2008.08.002 Martin, S. A. M., Blaney, S. C., Houlihan, D. F., & Secombes, C. J. (2006). Transcriptome response following administration of a live bacterial vaccine in Atlantic salmon (Salmo salar). Molecular Immunology, 43(11), 1900-1911. doi: 10.1016/j.molimm.2005.10.007 Martinez, D., Challacombe, J., Morgenstern, I., Hibbett, D., Schmoll, M., Kubicek, C. P., . . . Cullen, D. (2009). Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc Natl Acad Sci U S A, 106(6), 19541959. doi: 10.1073/pnas.0809575106 Mattila, T. M., Bechsgaard, J. S., Hansen, T. T., Schierup, M. H., & Bilde, T. (2012). Orthologous genes identified by transcriptome sequencing in the spider genus Stegodyphus. BMC genomics, 13. doi: 10.1186/1471-2164-13-70 Matz, C., Moreno, A. M., Alhede, M., Manefield, M., Hauser, A. R., Givskov, M., & Kjelleberg, S. (2008). Pseudomonas aeruginosa uses type III secretion system to kill biofilm-associated amoebae. Isme Journal, 2(8), 843-852. doi: 10.1038/ismej.2008.47 Matzkin, L. M. (2012). Population transcriptomics of cactus host shifts in Drosophila mojavensis. Molecular Ecology, 21(10), 2428-2439. doi: 10.1111/j.1365-294X.2012.05549.x Maughan, H., Birky, C. W., Jr., & Nicholson, W. L. (2009). Transcriptome Divergence and the Loss of Plasticity in Bacillus subtilis after 6,000 Generations of Evolution under Relaxed Selection for Sporulation. Journal of Bacteriology, 191(1), 428-433. doi: 10.1128/jb.01234-08 Mauter, M., Fait, A., Elimelech, M., & Herzberg, M. (2013). Surface Cell Density Effects on Escherichia coli Gene Expression during Cell Attachment.Environmental science & technology, 47(12), 62236230. Mavarez, J., Audet, C., & Bernatchez, L. (2009). Major disruption of gene expression in hybrids between young sympatric anadromous and resident populations of brook charr (Salvelinus fontinalis Mitchill). J Evol Biol, 22(8), 1708-1720. doi: 10.1111/j.1420-9101.2009.01785.x 38 McCoy, R. C., Garud, N. R., Kelley, J. L., Boggs, C. L., & Petrov, D. A. (2014). Genomic inference accurately predicts the timing and severity of a recent bottleneck in a nonmodel insect population. Molecular Ecology, 23(1), 136-150. doi: 10.1111/mec.12591 McDonnell, C. M., Alaux, C., Parrinello, H., Desvignes, J.-P., Crauser, D., Durbesson, E., . . . Le Conte, Y. (2013). Ecto- and endoparasite induce similar chemical and brain neurogenomic responses in the honey bee (Apis mellifera). Bmc Ecology, 13. doi: 10.1186/1472-6785-13-25 McKain, M. R., Wickett, N., Zhang, Y., Ayyampalayam, S., McCombie, W. R., Chase, M. W., . . . Leebens-Mack, J. (2012). PHYLOGENOMIC ANALYSIS OF TRANSCRIPTOME DATA ELUCIDATES CO-OCCURRENCE OF A PALEOPOLYPLOID EVENT AND THE ORIGIN OF BIMODAL KARYOTYPES IN AGAVOIDEAE (ASPARAGACEAE). American Journal of Botany, 99(2), 397-406. doi: 10.3732/ajb.1100537 Meier, K., Hansen, M. M., Normandeau, E., Mensberg, K.-L. D., Frydenberg, J., Larsen, P. F., . . . Bernatchez, L. (2014). Local adaptation at the transcriptome level in brown trout: evidence from early life history temperature genomic reaction norms. PloS one, 9(1), e85171. Mela, F., Fritsche, K., de Boer, W., van Veen, J. A., de Graaff, L. H., van den Berg, M., & Leveau, J. H. J. (2011). Dual transcriptional profiling of a bacterial/fungal confrontation: Collimonas fungivorans versus Aspergillus niger. Isme Journal, 5(9), 1494-1504. doi: 10.1038/ismej.2011.29 Meng, F., Braasch, I., Phillips, J. B., Lin, X., Titus, T., Zhang, C., & Postlethwait, J. H. (2013). Evolution of the Eye Transcriptome under Constant Darkness in Sinocyclocheilus Cavefish. Molecular biology and evolution, 30(7), 1527-1543. doi: 10.1093/molbev/mst079 Meyer, E., Aglyamova, G. V., & Matz, M. V. (2011). Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-Seq procedure. Molecular Ecology, 20(17), 3599-3616. doi: 10.1111/j.1365-294X.2011.05205.x Mezey, J. G., Nuzhdin, S. V., Ye, F., & Jones, C. D. (2008). Coordinated evolution of co-expressed gene clusters in the Drosophila transcriptome. Bmc Evolutionary Biology, 8. doi: 10.1186/1471-2148-8-2 Michely, S., Toulza, E., Subirana, L., John, U., Cognat, V., Maréchal-Drouard, L., . . . Piganeau, G. (2013). Evolution of codon usage in the smallest photosynthetic eukaryotes and their giant viruses. Genome Biology and Evolution, 5(5), 848-859. Milan, M., Coppe, A., Reinhardt, R., Cancela, L. M., Leite, R. B., Saavedra, C., . . . Bargelloni, L. (2011). Transcriptome sequencing and microarray development for the Manila clam, Ruditapes philippinarum: genomic tools for environmental monitoring. BMC genomics, 12. doi: 10.1186/14712164-12-234 Milan, M., Ferraresso, S., Ciofi, C., Chelazzi, G., Carrer, C., Ferrari, G., . . . Bargelloni, L. (2013). Exploring the effects of seasonality and chemical pollution on the hepatopancreas transcriptome of the Manila clam. Molecular Ecology, 22(8), 2157-2172. doi: 10.1111/mec.12257 Miller, H. C., Biggs, P. J., Voelckel, C., & Nelson, N. J. (2012). De novo sequence assembly and characterisation of a partial transcriptome for an evolutionarily distinct reptile, the tuatara (Sphenodon punctatus). BMC genomics, 13. doi: 10.1186/1471-2164-13-439 Mittapalli, O., Bai, X., Mamidala, P., Rajarapu, S. P., Bonello, P., & Herms, D. A. (2010). Tissue-Specific Transcriptomics of the Exotic Invasive Insect Pest Emerald Ash Borer (Agrilus planipennis). PloS one, 5(10). doi: 10.1371/journal.pone.0013708 Mok, B. W., Ribacke, U., Winter, G., Yip, B. H., Tan, C.-S., Fernandez, V., . . . Wahgren, M. (2007). Comparative transcriptornal analysis of isogenic Plasmodium falciparum clones of distinct antigenic and adhesive phenotypes. Molecular and Biochemical Parasitology, 151(2), 184-192. doi: 10.1016/j.molbiopara.2006.11.006 Moreno-Paz, M., Gómez, M. J., Arcas, A., & Parro, V. (2010). Environmental transcriptome analysis reveals physiological differences between biofilm and planktonic modes of life of the iron oxidizing bacteria Leptospirillum spp. in their natural microbial community. BMC genomics, 11(1), 404. 39 Moriwaki, T., Miyazawa, Y., & Takahashi, H. (2010). Transcriptome analysis of gene expression during the hydrotropic response in Arabidopsis seedlings. Environmental and Experimental Botany, 69(2), 148-157. doi: 10.1016/j.envexpbot.2010.03.013 Moya, A., Ganot, P., Furla, P., & Sabourault, C. (2012). The transcriptomic response to thermal stress is immediate, transient and potentiated by ultraviolet radiation in the sea anemone Anemonia viridis. Mol Ecol, 21(5), 1158-1174. doi: 10.1111/j.1365-294X.2012.05458.x Moyers, B. T., & Rieseberg, L. H. (2013). DIVERGENCE IN GENE EXPRESSION IS UNCOUPLED FROM DIVERGENCE IN CODING SEQUENCE IN A SECONDARILY WOODY SUNFLOWER. International Journal of Plant Sciences, 174(7), 1079-1089. doi: 10.1086/671197 Mukhopadhyay, A., He, Z. L., Alm, E. J., Arkin, A. P., Baidoo, E. E., Borglin, S. C., . . . Keasling, J. D. (2006). Salt stress in Desulfovibrio vulgaris Hildenborough: An integrated genomics a pproach. Journal of Bacteriology, 188(11), 4068-4078. doi: 10.1128/jb.01921-05 Nagano, A. J., Sato, Y., Mihara, M., Antonio, B. A., Motoyama, R., Itoh, H., . . . Izawa, T. (2012). Deciphering and prediction of transcriptome dynamics under fluctuating field conditions. Cell, 151(6), 1358-1369. doi: 10.1016/j.cell.2012.10.048 Nätt, D., Rubin, C.-J., Wright, D., Johnsson, M., Beltéky, J., Andersson, L., & Jensen, P. (2012). Heritable genome-wide variation of gene expression and promoter methylation between wild and domesticated chickens. BMC genomics, 13(1), 59. Navarro, A., Campos, B., Barata, C., & Pina, B. (2013). Transcriptomic seasonal variations in a natural population of zebra mussel (Dreissena polymorpha). Science of the Total Environment, 454, 482-489. doi: 10.1016/j.scitotenv.2013.03.048 Newton, J. R., Zenger, K. R., & Jerry, D. R. (2013). Next-generation transcriptome profiling reveals insights into genetic factors contributing to growth differences and temperature adaptation in Australian populations of barramundi (Lates calcarifer). Mar Genomics, 11, 45-52. doi: 10.1016/j.margen.2013.07.002 Nguyen, Q., Palfreyman, R. W., Chan, L. C., Reid, S., & Nielsen, L. K. (2012). Transcriptome sequencing of and microarray development for a Helicoverpa zea cell line to investigate in vitro insect cellbaculovirus interactions. PloS one, 7(5), e36324. Nikinmaa, M., McCairns, R. J. S., Nikinmaa, M. W., Vuori, K. A., Kanerva, M., Leinonen, T., . . . Leder, E. H. (2013). Transcription and redox enzyme activities: comparison of equilibrium and disequilibrium levels in the three-spined stickleback. Proceedings of the Royal Society B-Biological Sciences, 280(1755). doi: 10.1098/rspb.2012.2974 Nipitwattanaphon, M., Wang, J., Dijkstra, M. B., & Keller, L. (2013). A simple genetic basis for complex social behaviour mediates widespread gene expression differences. Molecular Ecology, 22(14), 37973813. doi: 10.1111/mec.12346 Niu, D., Wang, L., Sun, F., Liu, Z., & Li, J. (2013). Development of Molecular Resources for an Intertidal Clam, Sinonovacula constricta, Using 454 Transcriptome Sequencing. PloS one, 8(7). doi: 10.1371/journal.pone.0067456 Niu, S.-H., Li, Z.-X., Yuan, H.-W., Chen, X.-Y., Li, Y., & Li, W. (2013). Transcriptome characterisation of Pinus tabuliformis and evolution of genes in the Pinus phylogeny. BMC genomics, 14. doi: 10.1186/1471-2164-14-263 Norman, J. D., Ferguson, M. M., & Danzmann, R. G. (2014). Transcriptomics of salinity tolerance capacity in Arctic charr (Salvelinus alpinus): a comparison of gene expression profiles between divergent QTL genotypes. Physiological genomics, 46(4), 123-137. doi: 10.1152/physiolgenomics.00105.2013 Nota, B., van Straalen, N. M., Ylstra, B., & Roelofs, D. (2010). Gene expression microarray analysis of heat stress in the soil invertebrate Folsomia candida. Insect Molecular Biology, 19(3), 315-322. doi: 10.1111/j.1365-2583.2009.00990.x 40 Novo, M., Riesgo, A., Fernández-Guerra, A., & Giribet, G. (2013). Pheromone evolution, reproductive genes, and comparative transcriptomics in Mediterranean earthworms (Annelida, Oligochaeta, Hormogastridae). Molecular biology and evolution, mst074. Nowack, E. C. M., Vogel, H., Groth, M., Grossman, A. R., Melkonian, M., & Gloeckner, G. (2011). Endosymbiotic Gene Transfer and Transcriptional Regulation of Transferred Genes in Paulinella chromatophora. Molecular biology and evolution, 28(1), 407-422. doi: 10.1093/molbev/msq209 Nyberg, K. G., Conte, M. A., Kostyun, J. L., Forde, A., & Bely, A. E. (2012). Transcriptome characterization via 454 pyrosequencing of the annelid Pristina leidyi, an emerging model for studying the evolution of regeneration. BMC genomics, 13. doi: 10.1186/1471-2164-13-287 Nygren, K., Wallberg, A., Samils, N., Stajich, J. E., Townsend, J. P., Karlsson, M., & Johannesson, H. (2012). Analyses of expressed sequence tags in Neurospora reveal rapid evolution of genes associated with the early stages of sexual reproduction in fungi. Bmc Evolutionary Biology, 12(1), 229. O'Donnell, M. J., Todgham, A. E., Sewell, M. A., Hammond, L. M., Ruggiero, K., Fangue, N. A., . . . Hofmann, G. E. (2010). Ocean acidification alters skeletogenesis and gene expression in larval sea urchins. Marine Ecology Progress Series, 398, 157-171. doi: 10.3354/meps08346 Ometto, L., Cestaro, A., Ramasamy, S., Grassi, A., Revadi, S., Siozios, S., . . . Rota-Stabelli, O. (2013). Linking Genomics and Ecology to Investigate the Complex Evolution of an Invasive Drosophila Pest. Genome Biology and Evolution, 5(4), 745-757. doi: 10.1093/gbe/evt034 Ometto, L., Ross, K. G., Shoemaker, D., & Keller, L. (2012). Disruption of gene expression in hybrids of the fire ants Solenopsis invicta and Solenopsis richteri. Molecular Ecology, 21(10), 2488-2501. doi: 10.1111/j.1365-294X.2012.05544.x Osada, N., Kohn, M. H., & Wu, C.-I. (2006). Genomic inferences of the cis-regulatory nucleotide polymorphisms underlying gene expression differences between Drosophila melanogaster mating races. Molecular biology and evolution, 23(8), 1585-1591. doi: 10.1093/molbev/msl023 Overgaard, J., Sorensen, J. G., Jensen, L. T., Loeschcke, V., & Kristensen, T. N. (2010). Field tests reveal genetic variation for performance at low temperatures in Drosophila melanogaster. Functional Ecology, 24(1), 186-195. doi: 10.1111/j.1365-2435.2009.01615.x Ozawa, R., Nishimura, O., Yazawa, S., Muroi, A., Takabayashi, J., & Arimura, G.-i. (2012). Temperaturedependent, behavioural, and transcriptional variability of a tritrophic interaction consisting of bean, herbivorous mite, and predator. Molecular Ecology, 21(22), 5624-5635. doi: 10.1111/mec.12052 Padilla-Gamiño, J. L., Kelly, M. W., Evans, T. G., & Hofmann, G. E. (2013). Temperature and CO2 additively regulate physiology, morphology and genomic responses of larval sea urchins, Strongylocentrotus purpuratus. Proceedings of the Royal Society B: Biological Sciences, 280(1759). Pagarete, A., Le Corguille, G., Tiwari, B., Ogata, H., de Vargas, C., Wilson, W. H., & Allen, M. J. (2011). Unveiling the transcriptional features associated with coccolithovirus infection of natural Emiliania huxleyi blooms. Fems Microbiology Ecology, 78(3), 555-564. doi: 10.1111/j.1574-6941.2011.01191.x Page, R. B., Boley, M. A., Kump, D. K., & Voss, S. R. (2013). Genomics of a metamorphic timing QTL: met1 maps to a unique genomic position and regulates morph and species-specific patterns of brain transcription. Genome Biology and Evolution, 5(9), 1716-1730. Pallavicini, A., Canapa, A., Barucca, M., Alfoldi, J., Biscotti, M. A., Buonocore, F., . . . Scapigliati, G. (2013). Analysis of the transcriptome of the Indonesian coelacanth Latimeria menadoensis. BMC genomics, 14. doi: 10.1186/1471-2164-14-538 Palumbi, S. R., Barshis, D. J., Traylor-Knowles, N., & Bay, R. A. (2014). Mechanisms of reef coral resistance to future climate change. Science, 344(6186), 895-898. Pandey, S. P., Shahi, P., Gase, K., & Baldwin, I. T. (2008). Herbivory-induced changes in the small-RNA transcriptome and phytohormone signaling in Nicotiana attenuata. Proc Natl Acad Sci U S A, 105(12), 4559-4564. doi: 10.1073/pnas.0711363105 41 Parikh, A., Miranda, E. R., Katoh-Kurasawa, M., Fuller, D., Rot, G., Zagar, L., . . . Shaulsky, G. (2010). Conserved developmental transcriptomes in evolutionarily divergent species. Genome Biology, 11(3). doi: 10.1186/gb-2010-11-3-r35 Park, S., Keathley, D. E., & Han, K.-H. (2008). Transcriptional profiles of the annual growth cycle in Populus deltoides. Tree Physiology, 28(3), 321-329. Patell, S., Gu, M., Davenport, P., Givskov, M., Waite, R. D., & Welch, M. (2010). Comparative microarray analysis reveals that the core biofilm-associated transcriptome of Pseudomonas aeruginosa comprises relatively few genes. Environmental Microbiology Reports, 2(3), 440-448. doi: 10.1111/j.1758-2229.2010.00158.x Patzelt, D., Wang, H., Buchholz, I., Rohde, M., Groebe, L., Pradella, S., . . . Tomasch, J. (2013). You are what you talk: quorum sensing induces individual morphologies and cell division modes in Dinoroseobacter shibae. Isme Journal, 7(12), 2274-2286. doi: 10.1038/ismej.2013.107 Pauchet, Y., Wilkinson, P., van Munster, M., Augustin, S., Pauron, D., & Ffrench-Constant, R. H. (2009). Pyrosequencing of the midgut transcriptome of the poplar leaf beetle Chrysomela tremulae reveals new gene families in Coleoptera. Insect Biochemistry and Molecular Biology, 39(5-6), 403-413. doi: 10.1016/j.ibmb.2009.04.001 Pavey, S. A., Sutherland, B. J. G., Leong, J., Robb, A., von Schalburg, K., Hamon, T. R., . . . Nielsen, J. L. (2011). Ecological transcriptomics of lake-type and riverine sockeye salmon (Oncorhynchus nerka). Bmc Ecology, 11, 31-Article No.: 31. doi: 10.1186/1472-6785-11-31 Pellino, M., Hojsgaard, D., Schmutzer, T., Scholz, U., Hoerandl, E., Vogel, H., & Sharbel, T. F. (2013). Asexual genome evolution in the apomictic Ranunculus auricomus complex: examining the effects of hybridization and mutation accumulation. Molecular Ecology, 22(23), 5908-5921. doi: 10.1111/mec.12533 Pellizzari, C., Krasnov, A., Afanasyev, S., Vitulo, N., Franch, R., Pegolo, S., . . . Bargelloni, L. (2013). High mortality of juvenile gilthead sea bream (Sparus aurata) from photobacteriosis is associated with alternative macrophage activation and anti-inflammatory response: Results of gene expression profiling of early responses in the head kidney. Fish & Shellfish Immunology, 34(5), 1269-1278. doi: 10.1016/j.fsi.2013.02.007 Peng, X., Zha, W., He, R., Lu, T., Zhu, L., Han, B., & He, G. (2011). Pyrosequencing the midgut transcriptome of the brown planthopper, Nilaparvata lugens. Insect Molecular Biology, 20(6), 745762. doi: 10.1111/j.1365-2583.2011.01104.x Perdiguero, P., Barbero, M. d. C., Cervera, M. T., Collada, C., & Soto, A. (2013). Molecular response to water stress in two contrasting Mediterranean pines (< i> Pinus pinaster</i> and< i> Pinus pinea</i>). Plant Physiology and Biochemistry, 67, 199-208. Perez-Porro, A. R., Navarro-Gomez, D., Uriz, M. J., & Giribet, G. (2013). A NGS approach to the encrusting Mediterranean sponge Crella elegans (Porifera, Demospongiae, Poecilosclerida): transcriptome sequencing, characterization and overview of the gene expression along three life cycle stages. Mol Ecol Resour, 13(3), 494-509. doi: 10.1111/1755-0998.12085 Philipp, E. E. R., Kraemer, L., Melzner, F., Poustka, A. J., Thieme, S., Findeisen, U., . . . Rosenstiel, P. (2012). Massively Parallel RNA Sequencing Identifies a Complex Immune Gene Repertoire in the lophotrochozoan Mytilus edulis. PloS one, 7(3). doi: 10.1371/journal.pone.0033091 Philippe, R. N., Ralph, S. G., Mansfield, S. D., & Bohlmann, J. (2010). Transcriptome profiles of hybrid poplar (Populus trichocarpa x deltoides) reveal rapid changes in undamaged, systemic sink leaves after simulated feeding by forest tent caterpillar (Malacosoma disstria). New Phytologist, 188(3), 787802. doi: 10.1111/j.1469-8137.2010.03392.x Pierce, S. K., Fang, X., Schwartz, J. A., Jiang, X., Zhao, W., Curtis, N. E., . . . Wang, J. (2012). Transcriptomic Evidence for the Expression of Horizontally Transferred Algal Nuclear Genes in the 42 Photosynthetic Sea Slug, Elysia chlorotica. Molecular biology and evolution, 29(6), 1545-1556. doi: 10.1093/molbev/msr316 Pina, C., Pinto, F., Feijo, J. A., & Becker, J. D. (2005). Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant physiology, 138(2), 744-756. doi: 10.1104/pp.104.057935 Piveteau, P., Depret, G., Pivato, B., Garmyn, D., & Hartmann, A. (2011). Changes in Gene Expression during Adaptation of Listeria monocytogenes to the Soil Environment. PloS one, 6(9). doi: 10.1371/journal.pone.0024881 Place, S. P., Menge, B. A., & Hofmann, G. E. (2012). Transcriptome profiles link environmental variation and physiological response of Mytilus californianus between Pacific tides. Functional Ecology, 26(1), 144-155. doi: 10.1111/j.1365-2435.2011.01924.x Plett, J. M., Wilkins, O., Campbell, M. M., Ralph, S. G., & Regan, S. (2010). Endogenous overexpression of Populus MYB186 increases trichome density, improves insect pest resistance, and impacts plant growth. Plant Journal, 64(3), 419-432. doi: 10.1111/j.1365-313X.2010.04343.x Poelchau, M. F., Reynolds, J. A., Denlinger, D. L., Elsik, C. G., & Armbruster, P. A. (2011). A de novo transcriptome of the Asian tiger mosquito, Aedes albopictus, to identify candidate transcripts for diapause preparation. BMC genomics, 12. doi: 10.1186/1471-2164-12-619 Poelchau, M. F., Reynolds, J. A., Elsik, C. G., Denlinger, D. L., & Armbruster, P. A. (2013). RNA-Seq reveals early distinctions and late convergence of gene expression between diapause and quiescence in the Asian tiger mosquito, Aedes albopictus. Journal of Experimental Biology, 216(21), 4082-4090. doi: 10.1242/jeb.089508 Polato, N. R., Altman, N. S., & Baums, I. B. (2013). Variation in the transcriptional response of threatened coral larvae to elevated temperatures. Molecular Ecology, 22(5), 1366-1382. Polato, N. R., Vera, J. C., & Baums, I. B. (2011). Gene Discovery in the Threatened Elkhorn Coral: 454 Sequencing of the Acropora palmata Transcriptome. PloS one, 6(12). doi: 10.1371/journal.pone.0028634 Polato, N. R., Voolstra, C. R., Schnetzer, J., DeSalvo, M. K., Randall, C. J., Szmant, A. M., . . . Baums, I. B. (2010). Location-specific responses to thermal stress in larvae of the reef-building coral Montastraea faveolata. PloS one, 5(6), e11221. doi: 10.1371/journal.pone.0011221 Pontes, M. H., Babst, M., Lochhead, R., Oakeson, K., Smith, K., & Dale, C. (2008). Quorum Sensing Primes the Oxidative Stress Response in the Insect Endosymbiont, Sodalis glossinidius. PloS one, 3(10). doi: 10.1371/journal.pone.0003541 Porcelli, I., Reuter, M., Pearson, B. M., Wilhelm, T., & van Vliet, A. H. M. (2013). Parallel evolution of genome structure and transcriptional landscape in the Epsilonproteobacteria. BMC genomics, 14. doi: 10.1186/1471-2164-14-616 Portune, K. J., Voolstra, C. R., Medina, M., & Szmant, A. M. (2010). Development and heat stressinduced transcriptomic changes during embryogenesis of the scleractinian coral Acropora palmata. Mar Genomics, 3(1), 51-62. doi: 10.1016/j.margen.2010.03.002 Pravosudov, V. V., Roth, T. C., II, Forister, M. L., Ladage, L. D., Kramer, R., Schilkey, F., & Van Der Linden, A. M. (2013). Differential hippocampal gene expression is associated with climate-related natural variation in memory and the hippocampus in food-caching chickadees. Molecular Ecology, 22(2), 397-408. doi: 10.1111/mec.12146 Prince, E. G., Kirkland, D., & Demuth, J. P. (2010). Hyperexpression of the X Chromosome in Both Sexes Results in Extensive Female Bias of X-Linked Genes in the Flour Beetle. Genome Biology and Evolution, 2, 336-346. doi: 10.1093/gbe/evq024 43 Prosdocimi, F., Bittencourt, D., da Silva, F. R., Kirst, M., Motta, P. C., & Rech, E. L. (2011). Spinning Gland Transcriptomics from Two Main Clades of Spiders (Order: Araneae) - Insights on Their Molecular, Anatomical and Behavioral Evolution. PloS one, 6(6). doi: 10.1371/journal.pone.0021634 Provost, B., Jouan, V., Hilliou, F., Delobel, P., Bernardo, P., Ravallec, M., . . . Volkoff, A.-N. (2011). Lepidopteran transcriptome analysis following infection by phylogenetically unrelated polydnaviruses highlights differential and common responses. Insect Biochemistry and Molecular Biology, 41(8), 582-591. doi: 10.1016/j.ibmb.2011.03.010 Qiao, L., Yang, W., Fu, J., & Song, Z. (2013). Transcriptome Profile of the Green Odorous Frog (Odorrana margaretae). PloS one, 8(9). doi: 10.1371/journal.pone.0075211 Rajpurohit, S., Oliveira, C. A. C., Etges, W. J., & Gibbs, A. G. (2013). Functional genomic and phenotypic responses to desiccation in natural populations of a desert drosophilid. Molecular Ecology, 22(10), 2698-2715. doi: 10.1111/mec.12289 Renaut, S., & Bernatchez, L. (2011). Transcriptome-wide signature of hybrid breakdown associated with intrinsic reproductive isolation in lake whitefish species pairs (Coregonus spp. Salmonidae). Heredity, 106(6), 1003-1011. doi: 10.1038/hdy.2010.149 Renaut, S., Nolte, A. W., & Bernatchez, L. (2009). Gene Expression Divergence and Hybrid Misexpression between Lake Whitefish Species Pairs (Coregonus spp. Salmonidae). Molecular biology and evolution, 26(4), 925-936. doi: 10.1093/molbev/msp017 Renaut, S., Nolte, A. W., & Bernatchez, L. (2010). Mining transcriptome sequences towards identifying adaptive single nucleotide polymorphisms in lake whitefish species pairs (Coregonus spp. Salmonidae). Molecular Ecology, 19, 115-131. doi: 10.1111/j.1365-294X.2009.04477.x Renaut, S., Nolte, A. W., Rogers, S. M., Derome, N., & Bernatchez, L. (2011). SNP signatures of selection on standing genetic variation and their association with adaptive phenotypes along gradients of ecological speciation in lake whitefish species pairs (Coregonus spp.). Molecular Ecology, 20(3), 545-559. doi: 10.1111/j.1365-294X.2010.04952.x Renaut, S., Owens, G. L., & Rieseberg, L. H. (2014). Shared selective pressure and local genomic landscape lead to repeatable patterns of genomic divergence in sunflowers. Molecular Ecology, 23(2), 311-324. doi: 10.1111/mec.12600 Rendon-Anaya, M., Delaye, L., Possani, L. D., & Herrera-Estrella, A. (2012). Global Transcriptome Analysis of the Scorpion Centruroides noxius: New Toxin Families and Evolutionary Insights from an Ancestral Scorpion Species. PloS one, 7(8). doi: 10.1371/journal.pone.0043331 Rensink, W. A., Lee, Y., Liu, J., Iobst, S., Ouyang, S., & Buell, C. R. (2005). Comparative analyses of six solanaceous transcriptomes reveal a high degree of sequence conservation and species-specific transcripts. BMC genomics, 6. doi: 10.1186/1471-2164-6-124 Rey, S., Boltana, S., Vargas, R., Roher, N., & MacKenzie, S. (2013). Combining animal personalities with transcriptomics resolves individual variation within a wild-type zebrafish population and identifies underpinning molecular differences in brain function. Molecular Ecology, 22(24), 6100-6115. doi: 10.1111/mec.12556 Ribeiro, J. M. C., Labruna, M. B., Mans, B. J., Maruyama, S. R., Francischetti, I. M. B., Barizon, G. C., & de Miranda Santos, I. K. F. (2012). The sialotranscriptome of Antricola delacruzi female ticks is compatible with non-hematophagous behavior and an alternative source of food. Insect Biochemistry and Molecular Biology, 42(5), 332-342. doi: 10.1016/j.ibmb.2012.01.003 Richards, C. L., Rosas, U., Banta, J., Bhambhra, N., & Purugganan, M. D. (2012). Genome-wide patterns of Arabidopsis gene expression in nature. PLoS genetics, 8(4), e1002662. Richier, S., Rodriguez-Lanetty, M., Schnitzler, C. E., & Weis, V. M. (2008). Response of the symbiotic cnidarian Anthopleura elegantissima transcriptome to temperature and UV increase. Comparative Biochemistry and Physiology D-Genomics & Proteomics, 3(4), 283-289. doi: 10.1016/j.cbd.2008.08.001 44 Riley, R., Charron, P., Idnurm, A., Farinelli, L., Dalpe, Y., Martin, F., & Corradi, N. (2014). Extreme diversification of the mating type-high-mobility group (MATA-HMG) gene family in a plantassociated arbuscular mycorrhizal fungus. New Phytologist, 201(1), 254-268. doi: 10.1111/nph.12462 Rinker, D. C., Zhou, X., Pitts, R. J., Rokas, A., & Zwiebel, L. J. (2013). Antennal transcriptome profiles of anopheline mosquitoes reveal human host olfactory specialization in Anopheles gambiae. BMC genomics, 14, 1-14. Roberge, C., Normandeau, E., Einum, S., Guderley, H., & Bernatchez, L. (2008). Genetic consequences of interbreeding between farmed and wild Atlantic salmon: insights from the transcriptome. Molecular Ecology, 17(1), 314-324. doi: 10.1111/j.1365-294X.2007.03438.x Roberge, C., Guderley, H., & Bernatchez, L. (2007). Genomewide identification of genes under directional selection: Gene transcription Q(ST) scan in diverging Atlantic salmon subpopulations. Genetics, 177(2), 1011-1022. doi: 10.1534/genetics.107.073759 Robertson, L. S., & Cornman, R. S. (2014). Transcriptome resources for the frogs Lithobates clamitans and Pseudacris regilla, emphasizing antimicrobial peptides and conserved loci for phylogenetics. Mol Ecol Resour, 14(1), 178-183. doi: 10.1111/1755-0998.12164 Rodgers-Melnick, E., Mane, S. P., Dharmawardhana, P., Slavov, G. T., Crasta, O. R., Strauss, S. H., . . . DiFazio, S. P. (2012). Contrasting patterns of evolution following whole genome versus tandem duplication events in Populus. Genome Research, 22(1), 95-105. Rodriguez-Lanetty, M., Phillips, W. S., & Weis, V. M. (2006). Transcriptome analysis of a cnidariandinoflagellate mutualism reveals complex modulation of host gene expression. BMC genomics, 7. doi: 10.1186/1471-2164-7-23 Roeding, F., Borner, J., Kube, M., Klages, S., Reinhardt, R., & Burmester, T. (2009). A 454 sequencing approach for large scale phylogenomic analysis of the common emperor scorpion (Pandinus imperator). Molecular phylogenetics and evolution, 53(3), 826-834. doi: 10.1016/j.ympev.2009.08.014 Rokitta, S. D., de Nooijer, L. J., Trimborn, S., de Vargas, C., Rost, B., & John, U. (2011). Transcriptome analyses reveal differential gene expression patterns between the life-cycle stages of emiliania huxleyi (haptophyta) and reflect specialization to different ecological niches. Journal of Phycology, 47(4), 829-838. Ronges, D., Walsh, J. P., Sinclair, B. J., & Stillman, J. H. (2012). Changes in extreme cold tolerance, membrane composition and cardiac transcriptome during the first day of thermal acclimation in the porcelain crab Petrolisthes cinctipes. Journal of Experimental Biology, 215(11), 1824-1836. doi: 10.1242/jeb.069658 Rosenblum, E. B., Poorten, T. J., Settles, M., & Murdoch, G. K. (2012). Only skin deep: shared genetic response to the deadly chytrid fungus in susceptible frog species. Molecular Ecology, 21(13), 31103120. doi: 10.1111/j.1365-294X.2012.05481.x Salcedo, T., Upadhyay, R. J., Nagasaki, K., & Bhattacharya, D. (2012). Dozens of Toxin-Related Genes Are Expressed in a Nontoxic Strain of the Dinoflagellate Heterocapsa circularisquama. Molecular biology and evolution, 29(6), 1503-1506. doi: 10.1093/molbev/mss007 Salem, M., Kenney, P. B., Rexroad, C. E., III, & Yao, J. (2006). Microarray gene expression analysis in atrophying rainbow trout muscle: a unique nonmammalian muscle degradation model. Physiological genomics, 28(1), 33-45. doi: 10.1152/physiolgenomics.00114.2006 Samils, N., Gioti, A., Karlsson, M., Sun, Y., Kasuga, T., Bastiaans, E., . . . Johannesson, H. (2013). Sexlinked transcriptional divergence in the hermaphrodite fungus Neurospora tetrasperma. Proceedings of the Royal Society B: Biological Sciences, 280(1764). Sanogo, Y. O., Band, M., Blatti, C., Sinha, S., & Bell, A. M. (2012). Transcriptional regulation of brain gene expression in response to a territorial intrusion. Proceedings of the Royal Society B-Biological Sciences, 279(1749), 4929-4938. doi: 10.1098/rspb.2012.2087 45 Sanogo, Y. O., Hankison, S., Band, M., Obregon, A., & Bell, A. M. (2011). Brain Transcriptomic Response of Threespine Sticklebacks to Cues of a Predator. Brain Behavior and Evolution, 77(4), 270-285. doi: 10.1159/000328221 Santos, E. M., Kille, P., Workman, V. L., Paull, G. C., & Tyler, C. R. (2008). Sexually dimorphic gene expression in the brains of mature zebrafish. Comparative Biochemistry and Physiology a-Molecular & Integrative Physiology, 149(3), 314-324. doi: 10.1016/j.cbpa.2008.01.010 Sass, A. M., Schmerk, C., Agnoli, K., Norville, P. J., Eberl, L., Valvano, M. A., & Mahenthiralingam, E. (2013). The unexpected discovery of a novel low-oxygen-activated locus for the anoxic persistence of Burkholderia cenocepacia. Isme Journal, 7(8), 1568-1581. doi: 10.1038/ismej.2013.36 Sauvage, C., Derôme, N., Normandeau, E., Cyr, J. S.-., Audet, C., & Bernatchez, L. (2010). Fast transcriptional responses to domestication in the brook charr Salvelinus fontinalis. Genetics, 185(1), 105-112. Schmidt, H., Greshake, B., Feldmeyer, B., Hankeln, T., & Pfenninger, M. (2013). Genomic basis of ecological niche divergence among cryptic sister species of non-biting midges. BMC genomics, 14. doi: 10.1186/1471-2164-14-384 Schnitzler, C. E., & Weis, V. M. (2010). Coral larvae exhibit few measurable transcriptional changes during the onset of coral-dinoflagellate endosymbiosis. Mar Genomics, 3(2), 107-116. doi: 10.1016/j.margen.2010.08.002 Schoville, S. D., Barreto, F. S., Moy, G. W., Wolff, A., & Burton, R. S. (2012). Investigating the molecular basis of local adaptation to thermal stress: population differences in gene expression across the transcriptome of the copepod Tigriopus californicus. Bmc Evolutionary Biology, 12. doi: 10.1186/1471-2148-12-170 Schramm, F., Ganguli, A., Kiehlmann, E., Englich, G., Walch, D., & von Koskull-Doring, P. (2006). The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Molecular Biology, 60(5), 759-772. doi: 10.1007/s11103-0055750-x Schwartz, J. A., Curtis, N. E., & Pierce, S. K. (2010). Using Algal Transcriptome Sequences to Identify Transferred Genes in the Sea Slug, Elysia chlorotica. Evolutionary Biology, 37(1), 29-37. doi: 10.1007/s11692-010-9079-2 Schwartz, T. S., Tae, H., Yang, Y., Mockaitis, K., Van Hemert, J. L., Proulx, S. R., . . . Bronikowski, A. M. (2010). A garter snake transcriptome: pyrosequencing, de novo assembly, and sex-specific differences. BMC genomics, 11. doi: 10.1186/1471-2164-11-694 Schweizer, F., Bodenhausen, N., Lassueur, S., Masclaux, F. G., & Reymond, P. (2013). Differential contribution of transcription factors to Arabidopsis thaliana defense against Spodoptera littoralis. Frontiers in Plant Science, 4. doi: 10.3389/fpls.2013.00013 Scully, E. D., Hoover, K., Carlson, J. E., Tien, M., & Geib, S. M. (2013). Midgut transcriptome profiling of Anoplophora glabripennis, a lignocellulose degrading cerambycid beetle. BMC genomics, 14. doi: 10.1186/1471-2164-14-850 Seeb, J. E., Pascal, C. E., Grau, E. D., Seeb, L. W., Templin, W. D., Harkins, T., & Roberts, S. B. (2011). Transcriptome sequencing and high-resolution melt analysis advance single nucleotide polymorphism discovery in duplicated salmonids. Mol Ecol Resour, 11(2), 335-348. doi: 10.1111/j.17550998.2010.02936.x Sellin Jeffries, M. K., Mehinto, A. C., Carter, B. J., Denslow, N. D., & Kolok, A. S. (2012). Taking microarrays to the field: differential hepatic gene expression of caged fathead minnows from Nebraska watersheds. Environmental Science & Technology, 46(3), 1877-1885. Sen, R., Raychoudhury, R., Cai, Y., Sun, Y., Lietze, V.-U., Boucias, D. G., & Scharf, M. E. (2013). Differential impacts of juvenile hormone, soldier head extract and alternate caste phenotypes on host 46 and symbiont transcriptome composition in the gut of the termite Reticulitermes flavipes. BMC genomics, 14, 1-18. Seo, S., Gomi, K., Kaku, H., Abe, H., Seto, H., Nakatsu, S., . . . Ohashi, Y. (2012). Identification of natural diterpenes that inhibit bacterial wilt disease in tobacco, tomato and Arabidopsis. Plant Cell Physiol, 53(8), 1432-1444. doi: 10.1093/pcp/pcs085 Seto, Y., & Tamura, K. (2013). Extensive Differences in Antifungal Immune Response in Two Drosophila Species Revealed by Comparative Transcriptome Analysis. International Journal of Genomics. doi: 10.1155/2013/542139 Shaw, T. I., Srivastava, A., Chou, W.-C., Liu, L., Hawkinson, A., Glenn, T. C., . . . Schountz, T. (2012). Transcriptome Sequencing and Annotation for the Jamaican Fruit Bat (Artibeus jamaicensis). PloS one, 7(11). doi: 10.1371/journal.pone.0048472 Shi, Y., Yan, X., Zhao, P., Yin, H., Zhao, X., Xiao, H., . . . Ma, X.-F. (2013). Transcriptomic Analysis of a Tertiary Relict Plant, Extreme Xerophyte Reaumuria soongorica to Identify Genes Related to Drought Adaptation. PloS one, 8(5). doi: 10.1371/journal.pone.0063993 Shiao, M.-S., Chang, A. Y.-F., Liao, B.-Y., Ching, Y.-H., Lu, M.-Y. J., Chen, S. M., & Li, W.-H. (2012). Transcriptomes of Mouse Olfactory Epithelium Reveal Sexual Differences in Odorant Detection. Genome Biology and Evolution, 4(5), 703-712. doi: 10.1093/gbe/evs039 Shin, S. C., Kim, S. J., Lee, J. K., Ahn, D. H., Kim, M. G., Lee, H., . . . Park, H. (2012). Transcriptomics and Comparative Analysis of Three Antarctic Notothenioid Fishes. PloS one, 7(8). doi: 10.1371/journal.pone.0043762 Sikhakolli, U. R., Lopez-Giraldez, F., Li, N., Common, R., Townsend, J. P., & Trail, F. (2012). Transcriptome analyses during fruiting body formation in Fusarium graminearum and Fusarium verticillioides reflect species life history and ecology. Fungal Genetics and Biology, 49(8), 663-673. doi: 10.1016/j.fgb.2012.05.009 Sinha, A., Sommer, R. J., & Dieterich, C. (2012). Divergent gene expression in the conserved dauer stage of the nematodes Pristionchus pacificus and Caenorhabditis elegans. BMC genomics, 13. doi: 10.1186/1471-2164-13-254 Sloan, D. B., Keller, S. R., Berardi, A. E., Sanderson, B. J., Karpovich, J. F., & Taylor, D. R. (2012). De novo transcriptome assembly and polymorphism detection in the flowering plant Silene vulgaris (Caryophyllaceae). Mol Ecol Resour, 12(2), 333-343. doi: 10.1111/j.1755-0998.2011.03079.x Small, C. M., Carney, G. E., Mo, Q., Vannucci, M., & Jones, A. G. (2009). A microarray analysis of sexand gonad-biased gene expression in the zebrafish: Evidence for masculinization of the transcriptome. BMC genomics, 10. doi: 10.1186/1471-2164-10-579 Small, C. M., Harlin-Cognato, A. D., & Jones, A. G. (2013). Functional similarity and molecular divergence of a novel reproductive transcriptome in two male-pregnant Syngnathus pipefish species. Ecology and Evolution, 3(12), 4092-4108. doi: 10.1002/ece3.763 Smith, G., Fang, Y., Liu, X., Kenny, J., Cossins, A. R., de Oliveira, C. C., . . . Ritchie, M. G. (2013). TRANSCRIPTOME-WIDE EXPRESSION VARIATION ASSOCIATED WITH ENVIRONMENTAL PLASTICITY AND MATING SUCCESS IN CACTOPHILIC DROSOPHILA MOJAVENSIS. Evolution, 67(7), 1950-1963. doi: 10.1111/evo.12082 Smith, S., Bernatchez, L., & Beheregaray, L. B. (2013). RNA-seq analysis reveals extensive transcriptional plasticity to temperature stress in a freshwater fish species. BMC genomics, 14, 1-12. Sneddon, L. U., Margareto, J., & Cossins, A. R. (2005). The use of transcriptomics to address questions in behaviour: Production of a suppression subtractive hybridisation library from dominance hierarchies of rainbow trout. Physiological and Biochemical Zoology, 78(5), 695-705. doi: 10.1086/432141 47 Snell-rood, e. C., cash, a., han, m. V., kijimoto, t., andrews, j., & moczek, a. P. (2011). Developmental decoupling of alternative phenotypes: insights from the transcriptomes of horn-polyphenic beetles. Evolution, 65(1), 231-245. doi: 10.1111/j.1558-5646.2010.01106.x Soos, V., Sebestyen, E., Posta, M., Kohout, L., Light, M. E., Van Staden, J., & Balazs, E. (2012). Molecular aspects of the antagonistic interaction of smoke-derived butenolides on the germination process of Grand Rapids lettuce (Lactuca sativa) achenes. New Phytologist, 196(4), 1060-1073. doi: 10.1111/j.1469-8137.2012.04358.x Spiess, N., Oufir, M., Matusikova, I., Stierschneider, M., Kopecky, D., Homolka, A., . . . Wilhelm, E. (2012). Ecophysiological and transcriptomic responses of oak (Quercus robur) to long-term drought exposure and rewatering. Environmental and Experimental Botany, 77, 117-126. doi: 10.1016/j.envexpbot.2011.11.010 Srinivasan, J., Dillman, A. R., Macchietto, M. G., Heikkinen, L., Lakso, M., Fracchia, K. M., . . . Sternberg, P. W. (2013). The Draft Genome and Transcriptome of Panagrellus redivivus Are Shaped by the Harsh Demands of a Free-Living Lifestyle. Genetics, 193(4), 1279-+. doi: 10.1534/genetics.112.148809 St-Cyr, J., Derome, N., & Bernatchez, L. (2008). The transcriptomics of life-history trade-offs in whitefish species pairs (Coregonus sp.). Molecular Ecology, 17(7), 1850-1870. doi: 10.1111/j.1365294X.2008.03696.x Stillman, J. H., & Tagmount, A. (2009). Seasonal and latitudinal acclimatization of cardiac transcriptome responses to thermal stress in porcelain crabs, Petrolisthes cinctipes. Mol Ecol, 18(20), 4206-4226. doi: 10.1111/j.1365-294X.2009.04354.x Stoll, S., Feldhaar, H., & Gross, R. (2009). Transcriptional profiling of the endosymbiont Blochmannia floridanus during different developmental stages of its holometabolous ant host. Environmental Microbiology, 11(4), 877-888. doi: 10.1111/j.1462-2920.2008.01808.x Su, C.-l., Chen, W.-C., Lee, A.-Y., Chen, C.-Y., Chang, Y.-C. A., Chao, Y.-T., & Shih, M.-C. (2013). A Modified ABCDE Model of Flowering in Orchids Based on Gene Expression Profiling Studies of the Moth Orchid Phalaenopsis aphrodite. PloS one, 8(11). doi: 10.1371/journal.pone.0080462 Sun, J., Wang, M., Wang, H., Zhang, H., Zhang, X., Thiyagarajan, V., . . . Qiu, J. W. (2012). De novo assembly of the transcriptome of an invasive snail and its multiple ecological applications. Mol Ecol Resour, 12(6), 1133-1144. doi: 10.1111/1755-0998.12014 Sussarellu, R., Fabioux, C., Le Moullac, G., Fleury, E., & Moraga, D. (2010). Transcriptomic response of the Pacific oyster Crassostrea gigas to hypoxia. Mar Genomics, 3(3-4), 133-143. doi: 10.1016/j.margen.2010.08.005 Sutherland, B. J. G., Jantzen, S. G., Yasuike, M., Sanderson, D. S., Koop, B. F., & Jones, S. R. M. (2012). Transcriptomics of coping strategies in free-swimming Lepeophtheirus salmonis (Copepoda) larvae responding to abiotic stress. Molecular Ecology, 21(24), 6000-6014. doi: 10.1111/mec.12072 Szoevenyi, P., Rensing, S. A., Lang, D., Wray, G. A., & Shaw, A. J. (2011). Generation-Biased Gene Expression in a Bryophyte Model System. Molecular biology and evolution, 28(1), 803-812. doi: 10.1093/molbev/msq254 Szoevenyi, P., Ricca, M., Hock, Z., Shaw, J. A., Shimizu, K. K., & Wagner, A. (2013). Selection Is No More Efficient in Haploid than in Diploid Life Stages of an Angiosperm and a Moss. Molecular biology and evolution, 30(8), 1929-1939. doi: 10.1093/molbev/mst095 Tatematsu, K., Ward, S., Leyser, O., Kamiya, Y., & Nambara, E. (2005). Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in arabidopsis. Plant physiology, 138(2), 757-766. doi: 10.1104/pp.104.057984 Taylor, G., Street, N. R., Tricker, P. J., Sjodin, A., Graham, L., Skogstrom, O., . . . Jansson, S. (2005). The transcriptome of Populus in elevated CO2. New Phytologist, 167(1), 143-154. doi: 10.1111/j.14698137.2005.01450.x 48 Teo, S.-S., Ho, C.-L., Teoh, S., Rahim, R. A., & Phang, S.-M. (2009). TRANSCRIPTOMIC ANALYSIS OF GRACILARIA CHANGII (RHODOPHYTA) IN RESPONSE TO HYPER- AND HYPOOSMOTIC STRESSES. Journal of Phycology, 45(5), 1093-1099. doi: 10.1111/j.15298817.2009.00724.x Teulier, L., Degletagne, C., Rey, B., Tornos, J., Keime, C., de Dinechin, M., . . . Duchamp, C. (2012). Selective upregulation of lipid metabolism in skeletal muscle of foraging juvenile king penguins: an integrative study. Proceedings of the Royal Society B-Biological Sciences, 279(1737), 2464-2472. doi: 10.1098/rspb.2011.2664 Thompson, A. W., Huang, K., Saito, M. A., & Chisholm, S. W. (2011). Transcriptome response of highand low-light-adapted Prochlorococcus strains to changing iron availability. Isme Journal, 5(10), 1580-1594. doi: 10.1038/ismej.2011.49 Timmerhaus, G., Krasnov, A., Takle, H., Afanasyev, S., Nilsen, P., Rode, M., & Jørgensen, S. M. (2012). Comparison of Atlantic salmon individuals with different outcomes of cardiomyopathy syndrome (CMS). BMC genomics, 13(1), 205. Timmermans, M. J. T. N., Roelofs, D., Nota, B., Ylstra, B., & Holmstrup, M. (2009). Sugar sweet springtails: on the transcriptional response of Folsomia candida (Collembola) to desiccation stress. Insect Molecular Biology, 18(6), 737-746. doi: 10.1111/j.1365-2583.2009.00916.x Timmins-Schiffman, E. B., Friedman, C. S., Metzger, D. C., White, S. J., & Roberts, S. B. (2013). Genomic resource development for shellfish of conservation concern. Mol Ecol Resour, 13(2), 295305. doi: 10.1111/1755-0998.12052 Tisch, D., & Schmoll, M. (2013). Targets of light signalling in Trichoderma reesei. BMC genomics, 14. doi: 10.1186/1471-2164-14-657 Tisserant, E., Da Silva, C., Kohler, A., Morin, E., Wincker, P., & Martin, F. (2011). Deep RNA sequencing improved the structural annotation of the Tuber melanosporum transcriptome. New Phytologist, 189(3), 883-891. doi: 10.1111/j.1469-8137.2010.03597.x Tisserant, E., Kohler, A., Dozolme-Seddas, P., Balestrini, R., Benabdellah, K., Colard, A., . . . Martin, F. (2012). The transcriptome of the arbuscular mycorrhizal fungus Glomus intraradices (DAOM 197198) reveals functional tradeoffs in an obligate symbiont. New Phytologist, 193(3), 755-769. doi: 10.1111/j.1469-8137.2011.03948.x Todgham, A. E., & Hofmann, G. E. (2009). Transcriptomic response of sea urchin larvae Strongylocentrotus purpuratus to CO2-driven seawater acidification. Journal of Experimental Biology, 212(16), 2579-2594. doi: 10.1242/jeb.032540 Toffano-Nioche, C., Nguyen, A. N., Kuchly, C., Ott, A., Gautheret, D., Bouloc, P., & Jacq, A. (2012). Transcriptomic profiling of the oyster pathogen Vibrio splendidus opens a window on the evolutionary dynamics of the small RNA repertoire in the Vibrio genus. Rna-a Publication of the Rna Society, 18(12), 2201-2219. doi: 10.1261/rna.033324.112 Tomasch, J., Gohl, R., Bunk, B., Diez, M. S., & Wagner-Doebler, I. (2011). Transcriptional response of the photoheterotrophic marine bacterium Dinoroseobacter shibae to changing light regimes. Isme Journal, 5(12), 1957-1968. doi: 10.1038/ismej.2011.68 Toth, A. L., Varala, K., Henshaw, M. T., Rodriguez-Zas, S. L., Hudson, M. E., & Robinson, G. E. (2010). Brain transcriptomic analysis in paper wasps identifies genes associated with behaviour across social insect lineages. Proceedings of the Royal Society B-Biological Sciences, 277(1691), 2139-2148. doi: 10.1098/rspb.2010.0090 Toullec, J.-Y., Corre, E., Bernay, B., Thorne, M. A. S., Cascella, K., Ollivaux, C., . . . Clark, M. S. (2013). Transcriptome and Peptidome Characterisation of the Main Neuropeptides and Peptidic Hormones of a Euphausiid: The Ice Krill, Euphausia crystallorophias. PloS one, 8(8). doi: 10.1371/journal.pone.0071609 49 Traeger, S., Altegoer, F., Freitag, M., Gabaldon, T., Kempken, F., Kumar, A., . . . Nowrousian, M. (2013). The Genome and Development-Dependent Transcriptomes of Pyronema confluens: A Window into Fungal Evolution. PLoS genetics, 9(9). doi: 10.1371/journal.pgen.1003820 Travers, S. E., Tang, Z., Caragea, D., Garrett, K. A., Hulbert, S. H., Leach, J. E., . . . Fay, P. A. (2010). Variation in gene expression of Andropogon gerardii in response to altered environmental conditions associated with climate change. Journal of Ecology, 98(2), 374-383. Traylor-Knowles, N., Granger, B. R., Lubinski, T. J., Parikh, J. R., Garamszegi, S., Xia, Y., . . . Finnerty, J. R. (2011). Production of a reference transcriptome and transcriptomic database (PocilloporaBase) for the cauliflower coral, Pocillopora damicornis. BMC genomics, 12. doi: 10.1186/1471-2164-12-585 Truebano, M., Burns, G., Thorne, M. A. S., Hillyard, G., Peck, L. S., Skibinski, D. O. F., & Clark, M. S. (2010). Transcriptional response to heat stress in the Antarctic bivalve Latemula elliptica. Journal of Experimental Marine Biology and Ecology, 391(1-2), 65-72. doi: 10.1016/j.jembe.2010.06.011 Tsagkogeorga, G., Turon, X., Galtier, N., Douzery, E. J. P., & Delsuc, F. (2010). Accelerated Evolutionary Rate of Housekeeping Genes in Tunicates. Journal of Molecular Evolution, 71(2), 153167. doi: 10.1007/s00239-010-9372-9 Tymchuk, W., O'Reilly, P., Bittman, J., Macdonald, D., & Schulte, P. (2010). Conservation genomics of Atlantic salmon: variation in gene expression between and within regions of the Bay of Fundy. Mol Ecol, 19(9), 1842-1859. doi: 10.1111/j.1365-294X.2010.04596.x Tzika, A. C., Helaers, R., Schramm, G., & Milinkovitch, M. C. (2011). Reptilian-transcriptome v1.0, a glimpse in the brain transcriptome of five divergent Sauropsida lineages and the phylogenetic position of turtles. EvoDevo, 2. doi: 10.1186/2041-9139-2-19 Uebbing, S., Kunstner, A., Makinen, H., & Ellegren, H. (2013). Transcriptome Sequencing Reveals the Character of Incomplete Dosage Compensation across Multiple Tissues in Flycatchers. Genome Biology and Evolution, 5(8), 1555-1566. doi: 10.1093/gbe/evt114 Unal, E., Bucklin, A., Lenz, P., & Towle, D. (2013). Gene expression of the marine copepod< i> Calanus finmarchicus</i>: Responses to small-scale environmental variation in the Gulf of Maine (NW Atlantic Ocean). Journal of Experimental Marine Biology and Ecology, 446, 76-85. Valles, S. M., Oi, D. H., Yu, F., Tan, X.-X., & Buss, E. A. (2012). Metatranscriptomics and Pyrosequencing Facilitate Discovery of Potential Viral Natural Enemies of the Invasive Caribbean Crazy Ant, Nylanderia pubens. PloS one, 7(2). doi: 10.1371/journal.pone.0031828 van de Mortel, J. E., de Vos, R. C. H., Dekkers, E., Pineda, A., Guillod, L., Bouwmeester, K., . . . Raaijmakers, J. M. (2012). Metabolic and Transcriptomic Changes Induced in Arabidopsis by the Rhizobacterium Pseudomonas fluorescens SS101. Plant physiology, 160(4), 2173-2188. doi: 10.1104/pp.112.207324 Van de Waal, D. B., John, U., Ziveri, P., Reichart, G.-J., Hoins, M., Sluijs, A., & Rost, B. (2013). Ocean Acidification Reduces Growth and Calcification in a Marine Dinoflagellate. PloS one, 8(6). doi: 10.1371/journal.pone.0065987 van Ommen Kloeke, A. E., van Gestel, C. A., Styrishave, B., Hansen, M., Ellers, J., & Roelofs, D. (2012). Molecular and life-history effects of a natural toxin on herbivorous and non-target soil arthropods. Ecotoxicology, 21(4), 1084-1093. Van Puyvelde, S., Cloots, L., Engelen, K., Das, F., Marchal, K., Vanderleyden, J., & Spaepen, S. (2011). Transcriptome Analysis of the Rhizosphere Bacterium Azospirillum brasilense Reveals an Extensive Auxin Response. Microbial Ecology, 61(4), 723-728. doi: 10.1007/s00248-011-9819-6 Vellichirammal, N. N., Zera, A. J., Schilder, R. J., Wehrkamp, C., Riethoven, J.-J. M., & Brisson, J. A. (2014). De Novo Transcriptome Assembly from Fat Body and Flight Muscles Transcripts to Identify Morph-Specific Gene Expression Profiles in Gryllus firmus. PloS one, 9(1). doi: 10.1371/journal.pone.0082129 50 Vences, M., Hauswaldt, J. S., Steinfartz, S., Rupp, O., Goesmann, A., Kuenzel, S., . . . Smirnov, N. A. (2013). Radically different phylogeographies and patterns of genetic variation in two European brown frogs, genus Rana. Molecular phylogenetics and evolution, 68(3), 657-670. doi: 10.1016/j.ympev.2013.04.014 Venier, P., De Pitta, C., Pallavicini, A., Marsano, F., Varotto, L., Romualdi, C., . . . Lanfranchi, G. (2006). Development of mussel mRNA profiling: Can gene expression trends reveal coastal water pollution? Mutation Research-Fundamental and Molecular Mechanisms of Mutagenesis, 602(1-2), 121-134. doi: 10.1016/j.mrfmmm.2006.08.007 Vera, J. C., Wheat, C. W., Fescemyer, H. W., Frilander, M. J., Crawford, D. L., Hanski, I., & Marden, J. H. (2008). Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol Ecol, 17(7), 1636-1647. doi: 10.1111/j.1365-294X.2008.03666.x Vercruysse, M., Fauvart, M., Jans, A., Beullens, S., Braeken, K., Cloots, L., . . . Michiels, J. (2011). Stress response regulators identified through genome-wide transcriptome analysis of the (p) ppGppdependent response in Rhizobium etli. Genome Biology, 12(2). doi: 10.1186/gb-2011-12-2-r17 Vermeulen, C. J., Sorensen, P., Gagalova, K. K., & Loeschcke, V. (2013). Transcriptomic analysis of inbreeding depression in cold-sensitive Drosophila melanogaster shows upregulation of the immune response. J Evol Biol, 26(9), 1890-1902. doi: 10.1111/jeb.12183 Verta, J. P., Landry, C. R., & Mackay, J. J. (2013). Are long-lived trees poised for evolutionary change? Single locus effects in the evolution of gene expression networks in spruce. Mol Ecol, 22(9), 23692379. doi: 10.1111/mec.12189 Vesala, L., Salminen, T., Laiho, A., Hoikkala, A., & Kankare, M. (2012). Cold tolerance and cold‐ induced modulation of gene expression in two Drosophila virilis group species with different distributions. Insect Molecular Biology, 21(1), 107-118. Vezzi, A., Campanaro, S., D'Angelo, M., Simonato, F., Vitulo, N., Lauro, F. M., . . . Valle, G. (2005). Life at depth: Photobacterium profundum genome sequence and expression analysis. Science, 307(5714), 1459-1461. doi: 10.1126/science.1103341 Vicoso, B., Emerson, J. J., Zektser, Y., Mahajan, S., & Bachtrog, D. (2013). Comparative Sex Chromosome Genomics in Snakes: Differentiation, Evolutionary Strata, and Lack of Global Dosage Compensation. PLoS Biology, 11(8). doi: 10.1371/journal.pbio.1001643 Vidal-Dupiol, J., Zoccola, D., Tambutte, E., Grunau, C., Cosseau, C., Smith, K. M., . . . Tambutte, S. (2013). Genes Related to Ion-Transport and Energy Production Are Upregulated in Response to CO2Driven pH Decrease in Corals: New Insights from Transcriptome Analysis. PloS one, 8(3). doi: 10.1371/journal.pone.0058652 Voelckel, C., & Baldwin, I. T. (2004). Herbivore-induced plant vaccination. Part II. Array-studies reveal the transience of herbivore-specific transcriptional imprints and a distinct imprint from stress combinations. Plant Journal, 38(4), 650-663. doi: 10.1111/j.1365-313X.2004.02077.x Volkers, R. J. M., Snoek, L. B., Hubar, C. J. v. H., Coopman, R., Chen, W., Yang, W., . . . Kammenga, J. E. (2013). Gene-environment and protein-degradation signatures characterize genomic and phenotypic diversity in wild Caenorhabditis elegans populations. BMC Biol, 11, 13pp.-13pp. von Reumont, B. M., Blanke, A., Richter, S., Alvarez, F., Bleidorn, C., & Jenner, R. A. (2014). The First Venomous Crustacean Revealed by Transcriptomics and Functional Morphology: Remipede Venom Glands Express a Unique Toxin Cocktail Dominated by Enzymes and a Neurotoxin. Molecular biology and evolution, 31(1), 48-58. doi: 10.1093/molbev/mst199 von Schalburg, K. R., Cooper, G. A., Yazawa, R., Davidson, W. S., & Koop, B. F. (2008). Microarray analysis reveals differences in expression of cell surface and extracellular matrix components during development of the trout ovary and testis. Comparative Biochemistry and Physiology D-Genomics & Proteomics, 3(1), 78-90. doi: 10.1016/j.cbd.2007.10.001 51 Vonk, F. J., Casewell, N. R., Henkel, C. V., Heimberg, A. M., Jansen, H. J., McCleary, R. J. R., . . . Richardson, M. K. (2013). The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc Natl Acad Sci U S A, 110(51), 20651-20656. doi: 10.1073/pnas.1314702110 Voolstra, C. R., Schwarz, J. A., Schnetzer, J., Sunagawa, S., Desalvo, M. K., Szmant, A. M., . . . Medina, M. (2009). The host transcriptome remains unaltered during the establishment of coral-algal symbioses. Molecular Ecology, 18(9), 1823-1833. doi: 10.1111/j.1365-294X.2009.04167.x Vornanen, M., Hassinen, M., Koskinen, H., & Krasnov, A. (2005). Steady-state effects of temperature acclimation on the transcriptome of the rainbow trout heart. American Journal of PhysiologyRegulatory Integrative and Comparative Physiology, 289(4), R1177-R1184. doi: 10.1152/ajpregu.00157.2005 Waegele, H., Deusch, O., Haendeler, K., Martin, R., Schmitt, V., Christa, G., . . . Martin, W. (2011). Transcriptomic Evidence That Longevity of Acquired Plastids in the Photosynthetic Slugs Elysia timida and Plakobranchus ocellatus Does Not Entail Lateral Transfer of Algal Nuclear Genes. Molecular biology and evolution, 28(1), 699-706. doi: 10.1093/molbev/msq239 Wang, H., Ma, Z., Cui, F., Wang, X., Guo, W., Lin, Z., . . . Kang, L. (2012). Parental phase status affects the cold hardiness of progeny eggs in locusts. Functional Ecology, 26(2), 379-389. doi: 10.1111/j.1365-2435.2011.01927.x Wang, M.-H., Marinotti, O., Vardo-Zalik, A., Boparai, R., & Yan, G. (2011). Genome-Wide Transcriptional Analysis of Genes Associated with Acute Desiccation Stress in Anopheles gambiae. PloS one, 6(10). doi: 10.1371/journal.pone.0026011 Wang, S., Hou, R., Bao, Z., Du, H., He, Y., Su, H., . . . Li, Y. (2013). Transcriptome sequencing of Zhikong scallop (Chlamys farreri) and comparative transcriptomic analysis with Yesso scallop (Patinopecten yessoensis). PloS one, 8(5), e63927. Wang, S., O'Brien, T. R., Pava-Ripoll, M., & St Leger, R. J. (2011). Local adaptation of an introduced transgenic insect fungal pathogen due to new beneficial mutations. Proc Natl Acad Sci U S A, 108(51), 20449-20454. doi: 10.1073/pnas.1113824108 Wang, X.-W., Luan, J.-B., Li, J.-M., Su, Y.-L., Xia, J., & Liu, S.-S. (2011). Transcriptome analysis and comparison reveal divergence between two invasive whitefly cryptic species. BMC genomics, 12. doi: 10.1186/1471-2164-12-458 Wang, X.-W., Zhao, Q.-Y., Luan, J.-B., Wang, Y.-J., Yan, G.-H., & Liu, S.-S. (2012). Analysis of a native whitefly transcriptome and its sequence divergence with two invasive whitefly species. BMC genomics, 13. doi: 10.1186/1471-2164-13-529 Wang, X., Fang, X., Yang, P., Jiang, X., Jiang, F., Zhao, D., . . . Kang, L. (2014). The locust genome provides insight into swarm formation and long-distance flight. Nature Communications, 5, 1-9. doi: 10.1038/ncomms3957 Wang, Z., Lopez-Giraldez, F., Lehr, N., Farré, M., Common, R., Trail, F., & Townsend, J. P. (2014). Global gene expression and focused knockout analysis reveals genes associated with fungal fruiting body development in Neurospora crassa. Eukaryotic Cell, 13(1), 154-169. Warr, E., Aguilar, R., Dong, Y., Mahairaki, V., & Dimopoulos, G. (2007). Spatial and sex-specific dissection of the Anopheles gambiae midgut transcriptome. BMC genomics, 8. doi: 10.1186/14712164-8-37 Wayne, M. L., Telonis-Scott, M., Bono, L. M., Harshman, L., Kopp, A., Nuzhdin, S. V., & McIntyre, L. M. (2007). Simpler mode of inheritance of transcriptional variation in male Drosophila melanogaster. Proc Natl Acad Sci U S A, 104(47), 18577-18582. doi: 10.1073/pnas.0705441104 Webster, L. M., Paterson, S., Mougeot, F., Martinez-Padilla, J., & Piertney, S. B. (2011). Transcriptomic response of red grouse to gastro-intestinal nematode parasites and testosterone: implications for population dynamics. Mol Ecol, 20(5), 920-931. doi: 10.1111/j.1365-294X.2010.04906.x 52 Wei, D.-D., Chen, E.-H., Ding, T.-B., Chen, S.-C., Dou, W., & Wang, J.-J. (2013). De Novo Assembly, Gene Annotation, and Marker Discovery in Stored-Product Pest Liposcelis entomophila (Enderlein) Using Transcriptome Sequences. PloS one, 8(11). doi: 10.1371/journal.pone.0080046 Wei, X., Zhang, X., Shen, D., Wang, H., Wu, Q., Lu, P., . . . Li, X. (2013). Transcriptome Analysis of Barbarea vulgaris Infested with Diamondback Moth (Plutella xylostella) Larvae. PloS one, 8(5). doi: 10.1371/journal.pone.0064481 Weiss, Y., Foret, S., Hayward, D. C., Ainsworth, T., King, R., Ball, E. E., & Miller, D. J. (2013). The acute transcriptional response of the coral Acropora millepora to immune challenge: expression of GiMAP/IAN genes links the innate immune responses of corals with those of mammals and plants. BMC genomics, 14, 1-13. Wellband, K. W., & Heath, D. D. (2013). The relative contribution of drift and selection to transcriptional divergence among Babine Lake tributary populations of juvenile rainbow trout. J Evol Biol, 26(11), 2497-2508. doi: 10.1111/jeb.12247 Wenzel, M. A., Webster, L. M. I., Paterson, S., Mougeot, F., Martinez-Padilla, J., & Piertney, S. B. (2013). A transcriptomic investigation of handicap models in sexual selection. Behavioral Ecology and Sociobiology, 67(2), 221-234. doi: 10.1007/s00265-012-1442-0 Werner, G. D. A., Gemmell, P., Grosser, S., Hamer, R., & Shimeld, S. M. (2013). Analysis of a deep transcriptome from the mantle tissue of Patella vulgata Linnaeus (Mollusca: Gastropoda: Patellidae) reveals candidate biomineralising genes. Marine Biotechnology, 15(2), 230-243. doi: 10.1007/s10126012-9481-0 Wertheim, B., Kraaijeveld, A. R., Hopkins, M. G., Boer, M. W., & Godfray, H. C. J. (2011). Functional genomics of the evolution of increased resistance to parasitism in Drosophila. Molecular Ecology, 20(5), 932-949. doi: 10.1111/j.1365-294X.2010.04911.x Wheat, C. W., Fescemyer, H. W., Kvist, J., Tas, E., Vera, J. C., Frilander, M. J., . . . Marden, J. H. (2011). Functional genomics of life history variation in a butterfly metapopulation. Mol Ecol, 20(9), 18131828. doi: 10.1111/j.1365-294X.2011.05062.x Whitehead, A., Dubansky, B., Bodinier, C., Garcia, T. I., Miles, S., Pilley, C., . . . Galvez, F. (2012). Genomic and physiological footprint of the Deepwater Horizon oil spill on resident marsh fishes. Proc Natl Acad Sci U S A, 109(50), 20298-20302. doi: 10.1073/pnas.1109545108 Whitehead, A., Galvez, F., Zhang, S., Williams, L. M., & Oleksiak, M. F. (2011). Functional Genomics of Physiological Plasticity and Local Adaptation in Killifish. Journal of Heredity, 102(5), 499-511. doi: 10.1093/jhered/esq077 Whitehead, A., Pilcher, W., Champlin, D., & Nacci, D. (2012). Common mechanism underlies repeated evolution of extreme pollution tolerance. Proceedings of the Royal Society B-Biological Sciences, 279(1728), 427-433. doi: 10.1098/rspb.2011.0847 Whitehead, A., Roach, J. L., Zhang, S., & Galvez, F. (2011). Genomic mechanisms of evolved physiological plasticity in killifish distributed along an environmental salinity gradient. Proc Natl Acad Sci U S A, 108(15), 6193-6198. doi: 10.1073/pnas.1017542108 Whitehead, A., Roach, J. L., Zhang, S., & Galvez, F. (2012). Salinity- and population-dependent genome regulatory response during osmotic acclimation in the killifish (Fundulus heteroclitus) gill. Journal of Experimental Biology, 215(8), 1293-1305. doi: 10.1242/jeb.062075 Whitehead, A., Triant, D. A., Champlin, D., & Nacci, D. (2010). Comparative transcriptomics implicates mechanisms of evolved pollution tolerance in a killifish population. Molecular Ecology, 19(23), 51865203. doi: 10.1111/j.1365-294X.2010.04829.x Whitehead, A., Zhang, S., Roach, J. L., & Galvez, F. (2013). Common functional targets of adaptive micro- and macro-evolutionary divergence in killifish. Molecular Ecology, 22(14), 3780-3796. doi: 10.1111/mec.12316 53 Whiteley, A. R., Derôme, N., Rogers, S. M., St-Cyr, J., Laroche, J., Labbe, A., & Bernatchez, L. (2008). The phenomics and eQTL mapping of brain transcriptomes regulating adaptive divergence in lake whitefish species pairs (Coregonus sp.). Genetics. Whiteman, N. K., Gloss, A. D., Sackton, T. B., Groen, S. C., Humphrey, P. T., Lapoint, R. T., . . . Pierce, N. E. (2012). Genes Involved in the Evolution of Herbivory by a Leaf-Mining, Drosophilid Fly. Genome Biology and Evolution, 4(9), 900-916. doi: 10.1093/gbe/evs063 Whittle, C. A., & Johannesson, H. (2013). Evolutionary dynamics of sex-biased genes in a hermaphrodite fungus. Molecular biology and evolution, 30(11), 2435-2446. Wier, A. M., Nyholm, S. V., Mandel, M. J., Massengo-Tiasse, R. P., Schaefer, A. L., Koroleva, I., . . . McFall-Ngai, M. J. (2010). Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci U S A, 107(5), 2259-2264. doi: 10.1073/pnas.0909712107 Williams, T. D., Diab, A., Ortega, F., Sabine, V. S., Godfrey, R. E., Falciani, F., . . . George, S. G. (2008). Transcriptomic responses of European flounder (Platichthys flesus) to model toxicants. Aquatic Toxicology, 90(2), 83-91. doi: 10.1016/j.aquatox.2008.07.019 Wisecaver, J. H., & Hackett, J. D. (2010). Transcriptome analysis reveals nuclear-encoded proteins for the maintenance of temporary plastids in the dinoflagellate Dinophysis acuminata. BMC genomics, 11. doi: 10.1186/1471-2164-11-366 Wolf, J. B., Bayer, T., Haubold, B., Schilhabel, M., Rosenstiel, P., & Tautz, D. (2010). Nucleotide divergence vs. gene expression differentiation: comparative transcriptome sequencing in natural isolates from the carrion crow and its hybrid zone with the hooded crow. Molecular Ecology, 19(s1), 162-175. Wong, E. S. W., Nicol, S., Warren, W. C., & Belov, K. (2013). Echidna Venom Gland Transcriptome Provides Insights into the Evolution of Monotreme Venom. PloS one, 8(11). doi: 10.1371/journal.pone.0079092 Woodard, S. H., Fischman, B. J., Venkat, A., Hudson, M. E., Varala, K., Cameron, S. A., . . . Robinson, G. E. (2011). Genes involved in convergent evolution of eusociality in bees. Proc Natl Acad Sci U S A, 108(18), 7472-7477. doi: 10.1073/pnas.1103457108 Wurmser, F., Ogereau, D., Mary-Huard, T., Loriod, B., Joly, D., & Montchamp-Moreau, C. (2011). Population transcriptomics: insights from Drosophila simulans, Drosophila sechellia and their hybrids. Genetica, 139(4), 465-477. doi: 10.1007/s10709-011-9566-0 Xi, Z., Gavotte, L., Xie, Y., & Dobson, S. L. (2008). Genome-wide analysis of the interaction between the endosymbiotic bacterium Wolbachia and its Drosophila host. BMC genomics, 9. doi: 10.1186/14712164-9-1 Xiang, L.-x., He, D., Dong, W.-r., Zhang, Y.-w., & Shao, J.-z. (2010). Deep sequencing-based transcriptome profiling analysis of bacteria-challenged Lateolabrax japonicus reveals insight into the immune-relevant genes in marine fish. BMC genomics, 11. doi: 10.1186/1471-2164-11-472 Xu, J., & James, R. (2012). Temperature stress affects the expression of immune response genes in the alfalfa leafcutting bee, Megachile rotundata. Insect Molecular Biology, 21(2), 269-280. Xu, J., Li, Q., Xu, L., Wang, S., Jiang, Y., Zhao, Z., . . . Xu, P. (2013). Gene expression changes leading extreme alkaline tolerance in Amur ide (Leuciscus waleckii) inhabiting soda lake. BMC genomics, 14(1), 682. Xu, Y., Zhou, W., Zhou, Y., Wu, J., & Zhou, X. (2012). Transcriptome and Comparative Gene Expression Analysis of Sogatella furcifera (Horvath) in Response to Southern Rice Black-Streaked Dwarf Virus. PloS one, 7(4). doi: 10.1371/journal.pone.0036238 54 Xue, J., Bao, Y.-Y., Li, B.-l., Cheng, Y.-B., Peng, Z.-Y., Liu, H., . . . Zhang, C.-X. (2010). Transcriptome Analysis of the Brown Planthopper Nilaparvata lugens. PloS one, 5(12). doi: 10.1371/journal.pone.0014233 Yampolsky, L. Y., Glazko, G. V., & Fry, J. D. (2012). Evolution of gene expression and expression plasticity in long-term experimental populations of Drosophila melanogaster maintained under constant and variable ethanol stress. Mol Ecol, 21(17), 4287-4299. doi: 10.1111/j.1365294X.2012.05697.x Yanai, I., Peshkin, L., Jorgensen, P., & Kirschner, M. W. (2011). Mapping Gene Expression in Two Xenopus Species: Evolutionary Constraints and Developmental Flexibility. Developmental Cell, 20(4), 483-496. doi: 10.1016/j.devcel.2011.03.015 Yang, I., Beszteri, S., Tillmann, U., Cembella, A., & John, U. (2011). Growth-and nutrient-dependent gene expression in the toxigenic marine dinoflagellate< i> Alexandrium minutum</i>. Harmful Algae, 12, 55-69. Yang, W., Qi, Y., Bi, K., & Fu, J. (2012). Toward understanding the genetic basis of adaptation to highelevation life in poikilothermic species: A comparative transcriptomic analysis of two ranid frogs, Rana chensinensis and R. kukunoris. BMC genomics, 13(1), 588. Yek, S. H., Boomsma, J. J., & Schiott, M. (2013). Differential gene expression in Acromyrmex leafcutting ants after challenges with two fungal pathogens. Molecular Ecology, 22(8), 2173-2187. doi: 10.1111/mec.12255 Ying, B.-W., Seno, S., Kaneko, F., Matsuda, H., & Yomo, T. (2013). Multilevel comparative analysis of the contributions of genome reduction and heat shock to the Escherichia coli transcriptome. BMC genomics, 14. doi: 10.1186/1471-2164-14-25 Yoo, M.-J., Chanderbali, A. S., Altman, N. S., Soltis, P. S., & Soltis, D. E. (2010). Evolutionary trends in the floral transcriptome: insights from one of the basalmost angiosperms, the water lily Nuphar advena (Nymphaeaceae). Plant Journal, 64(4), 687-698. doi: 10.1111/j.1365-313X.2010.04357.x Yoshida, M.-a., & Ogura, A. (2011). Genetic mechanisms involved in the evolution of the cephalopod camera eye revealed by transcriptomic and developmental studies. Bmc Evolutionary Biology, 11. doi: 10.1186/1471-2148-11-180 Yuan, J.-B., Zhang, X.-J., Liu, C.-Z., Wei, J.-K., Li, F.-H., & Xiang, J.-H. (2013). Horizontally transferred genes in the genome of Pacific white shrimp, Litopenaeus vannamei. Bmc Evolutionary Biology, 13. doi: 10.1186/1471-2148-13-165 Yuan, M., Chen, M., Zhang, W., Lu, W., Wang, J., Yang, M., . . . Lin, M. (2012). Genome Sequence and Transcriptome Analysis of the Radioresistant Bacterium Deinococcus gobiensis. Insights into the Extreme Environmental Adaptations. PloS one, 7(3). doi: 10.1371/journal.pone.0034458 Yudkovski, Y., Ramsak, A., Ausland, M., & Tom, M. (2010). Potential of the hepatic transcriptome expression profile of the striped seabream (Lithognathus mormyrus) as an environmental biomarker. Biomarkers, 15(7), 625-638. doi: 10.3109/1354750x.2010.510579 Zahn, L. M., Ma, X., Altman, N. S., Zhang, Q., Wall, P. K., Tian, D., . . . Ma, H. (2010). Comparative transcriptomics among floral organs of the basal eudicot Eschscholzia californica as reference for floral evolutionary developmental studies. Genome Biology, 11(10). doi: 10.1186/gb-2010-11-10-r101 Zakas, C., Schult, N., McHugh, D., Jones, K. L., & Wares, J. P. (2012). Transcriptome Analysis and SNP Development Can Resolve Population Differentiation of Streblospio benedicti, a Developmentally Dimorphic Marine Annelid. PloS one, 7(2). doi: 10.1371/journal.pone.0031613 Zhai, Y., Zhang, J., Sun, Z., Dong, X., He, Y., Kang, K., . . . Zhang, W. (2013). Proteomic and Transcriptomic Analyses of Fecundity in the Brown Planthopper Nilaparvata lugens (Stal). Journal of Proteome Research, 12(11), 5199-5212. doi: 10.1021/pr400561c 55 Zhang, L., Yan, H.-F., Wu, W., Yu, H., & Ge, X.-J. (2013). Comparative transcriptome analysis and marker development of two closely related Primrose species (Primula poissonii and Primula wilsonii). BMC genomics, 14. doi: 10.1186/1471-2164-14-329 Zhang, S., Zhang, Z., & Kang, L. (2012). Transcriptome response analysis of Arabidopsis thaliana to leafminer (Liriomyza huidobrensis). Bmc Plant Biology, 12. doi: 10.1186/1471-2229-12-234 Zhang, W., Culley, D. E., Nie, L., & Brockman, F. J. (2006). DNA microarray analysis of anaerobic Methanosarcina barkeri reveals responses to heat shock and air exposure. Journal of Industrial Microbiology & Biotechnology, 33(9), 784-790. doi: 10.1007/s10295-006-0114-3 Zhang, W., Kunte, K., & Kronforst, M. R. (2013). Genome-Wide Characterization of Adaptation and Speciation in Tiger Swallowtail Butterflies Using De Novo Transcriptome Assemblies. Genome Biology and Evolution, 5(6), 1233-1245. doi: 10.1093/gbe/evt090 Zhang, Y.-N., Jin, J.-Y., Jin, R., Xia, Y.-H., Zhou, J.-J., Deng, J.-Y., & Dong, S.-L. (2013). Differential Expression Patterns in Chemosensory and Non-Chemosensory Tissues of Putative Chemosensory Genes Identified by Transcriptome Analysis of Insect Pest the Purple Stem Borer Sesamia inferens (Walker). PloS one, 8(7). doi: 10.1371/journal.pone.0069715 Zhang, Z., Wang, Y., Wang, S., Liu, J., Warren, W., Mitreva, M., & Walter, R. B. (2011). Transcriptome analysis of female and male Xiphophorus maculatus Jp 163 A. PloS one, 6(4), e18379. Zhao, S., Shalchian-Tabrizi, K., & Klaveness, D. (2013). Sulcozoa revealed as a paraphyletic group in mitochondrial phylogenomics. Molecular phylogenetics and evolution, 69(3), 462-468. Zhao, Z., Wu, G., Wang, J., Liu, C., & Qiu, L. (2013). Next-Generation Sequencing-Based Transcriptome Analysis of Helicoverpa armigera Larvae Immune-Primed with Photorhabdus luminescens TT01. PloS one, 8(11). doi: 10.1371/journal.pone.0080146 Zheng, M., Tian, J., Yang, G., Zheng, L., Chen, G., Chen, J., & Wang, B. (2013). Transcriptome sequencing, annotation and expression analysis of Nannochloropsis sp at different growth phases. Gene, 523(2), 117-121. doi: 10.1016/j.gene.2013.04.005 Zheng, W., Peng, T., He, W., & Zhang, H. (2012). High-Throughput Sequencing to Reveal Genes Involved in Reproduction and Development in Bactrocera dorsalis (Diptera: Tephritidae). PloS one, 7(5). doi: 10.1371/journal.pone.0036463 Zhou, A., Baidoo, E., He, Z., Mukhopadhyay, A., Baumohl, J. K., Benke, P., . . . Zhou, J. (2013). Characterization of NaCl tolerance in Desulfovibrio vulgaris Hildenborough through experimental evolution. Isme Journal, 7(9), 1790-1802. doi: 10.1038/ismej.2013.60 Zhou, Z. C., Dong, Y., Sun, H. J., Yang, A. F., Chen, Z., Gao, S., . . . Wang, B. (2014). Transcriptome sequencing of sea cucumber (Apostichopus japonicus) and the identification of gene-associated markers. Mol Ecol Resour, 14(1), 127-138. doi: 10.1111/1755-0998.12147 Zhu, J.-Y., Li, Y.-H., Yang, S., & Li, Q.-W. (2013). De novo Assembly and Characterization of the Global Transcriptome for Rhyacionia leptotubula Using Illumina Paired-End Sequencing. PloS one, 8(11). doi: 10.1371/journal.pone.0081096 Zhu, J.-Y., Zhao, N., & Yang, B. (2012). Global Transcriptome Profiling of the Pine Shoot Beetle, Tomicus yunnanensis (Coleoptera: Scolytinae). PloS one, 7(2). doi: 10.1371/journal.pone.0032291 Zhu, L., Yan, Z., Feng, M., Peng, D., Guo, Y., Hu, X., . . . Sun, Y. (2014). Identification of sturgeon IgD bridges the evolutionary gap between elasmobranchs and teleosts. Developmental and Comparative Immunology, 42(2), 138-147. doi: 10.1016/j.dci.2013.08.020 56