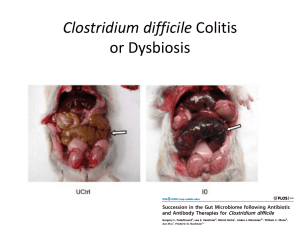
The NIH Human Microbiome Project (HMP)
advertisement

The NIH Human Microbiome Project (HMP) Prepared by Lita M. Proctor, NHGRI/NIH for 2013 ACVP Conference proceedings 081913 draft Abstract The human microbiome comprises the genes, gene products and genomes of the microbiota, which include archaea, bacteria, eukaryotic viruses, bacteriophage, fungi and protozoa, that inhabit the body. The NIH Human Microbiome Project (HMP) was established to create a map of the microbial communities of the human body in health and in disease and to evaluate the biological properties of the microbiome. As a community resource program, the HMP has produced or will produce reference microbial genome sequences, 16S ribosomal RNA gene sequences for microbial taxonomic analysis, metagenome sequences for microbial community analysis, as well microbial community gene expression, proteome and metabolome data from the microbiome. All of these data as well as associated computational tools and scientific approaches are being generated or developed to support growth of this emerging concept for human health. Introduction Throughout most of human history, we have felt at war with microbes. Bubonic plague, small pox, yellow fever, and typhoid are just a few important historical examples while modern day infectious diseases include malaria, tuberculosis, cholera and HIV/AIDS, to name a few. The scientific study of microbiology, which led to important discoveries such as Louis Pasteur’s “germ theory of disease”, grew out of society’s desire to conquer these pathogens and eradicate infectious disease. But a new view is emerging in which we recognize humans and microbes as a co-evolved system for the mutual benefit of both the host and the resident microbes. We now recognize that the human body is made up of about 10 times more microbial cells (~1014) than human cells (~1013). Further, there may be hundreds of times more microbial genes (i.e. millions) than human genes (i.e. 20,000-25,000) in this human+microbiome system, which is often thought of as a human ‘superorganism’, and it is these microbial communities and the way they interact with the human host which describe their role in our health. It is thought that infants are sterile in the womb and receive their first inoculum of microbes from the mother, which goes on to colonize the newborn and, through successional waves of different microbes, leads to the development of its own microbiome. The newborn relies on this maternal microbial inoculum as well as other environmental sources of microbes for microbial colonization of all exposed surfaces in and on the infant’s body (e.g. oral, nasal/airways, gut, urogenital, skin). This is a dynamic process in which microbial abundances increase from effectively zero at birth to over 6 orders of magnitude within just the first few weeks of life with wide swings in the microbial membership of these communities until the microbiota largely stabilize and become ‘adult-like’ in composition and numbers after approx. 3 years of life (Costello et al., 2009; Koenig et al., 2011). At the same time, it is thought that the newborn’s gut microbiota trigger development and maturation of the newborn’s immune system. Though there is still a great deal of active research needed to understand precisely what happens in this developmental process, it appears that the maturing immune system relies on the presence of specific microbial communities and especially the presence of specific bacteria early in the maturation process as the immunological trigger for identifying ‘self’ from ‘nonself’ (Hooper and Gordon, 2001; Round and Mazmanian, 2009). Much research is now examining the links 1 between these early life events of the microbiome and the immune system and the subsequent health status over a person’s lifetime. The NIH Human Microbiome Project (HMP) was created to provide tools, data and resources to enable research in this emerging concept in the biomedical field and for human health. The NIH Human Microbiome Project: A Community Resource The human microbiome encompasses the full complement of microbial genes, gene products, and genomes of the microbiota (which include bacteria, archaea, eukaryotic viruses, bacteriophage and eukaryotic microbes like fungi and protozoa) that call the human body home and interact with the human host to maintain host health. At thousands of species, millions of microbial genes and trillions of microbial cells, the global microbiome contributes to the health and maintenance of the human superorganism. Interest in this human+microbiome system has been motivated by simultaneous advances in sequencing technologies and in microbial ecology, by an ever increasing understanding that the human host and microbiota have co-evolved and that resident microbiota are intimately involved in the development and maintenance of the human immune system. To catalyze this research, the NIH launched an 8-year Human Microbiome Project (HMP) as a community resource program (http://commonfund.nih.gov/hmp/). There are two phases to the program, a first phase (2007-2012) to undertake a survey of the human microbiome and a second phase (2013-2015) to create a catalog of biological properties of the human microbiome. The HMP is 1) surveying the microbiomes across the bodies of a cohort of healthy adults to produce a reference dataset of baseline microbiomes, 2) developing a catalog of microbial genome sequences of reference strains and, 3) evaluating the characteristics of microbiomes associated with specific gastrointestinal tract, urogenital and skin diseases and 4) compiling a rich catalog of microbiome biological properties in association with host properties from cohorts with known diseases or health conditions. A description of the some of the key resources and the consortium activities in both phases of this program follow. HMP Reference Strain Microbial Genome Sequence Catalog The HMP has assembled a key reference dataset of microbial genome sequences collected from the major body regions of the human microbiome, primarily bacterial, although it also includes archaea, viruses, bacteriophage and eukaryotic microorganisms. The project's target catalog of 3,000 microbial genome sequences is intended as a reference for the interpretation of the 16S ribosomal RNA gene sequences, as well as a scaffold for assembly of metagenomic sequences determined from the microbial communities. A publication documenting the analysis of the first 178 microbial isolates was published (Nelson et al., 2010); just this subset of the catalog described over 550,000 predicted genes, 30,000 of which were novel. As of this writing, thousands of microbial strains have been sequenced or are in progress for the HMP reference strain catalog (http://www.hmpdacc-resources.org/hmp_catalog). Several hundred of these sequences are available in GenBank (http://www.ncbi.nlm.nih.gov/bioproject/28331), and a key subset of the cultures of the corresponding reference strains are available at the HMP Strain Repository in the ATCC/Biodefense and Emerging Infectious Diseases Research Repository (BEI) (http://www.beiresources.org/Collection/4/Human-Microbiome-Project.aspx). HMP Healthy Adult Cohort Study The second major resource of the HMP is the largest study to date of the microbiomes of five major regions of the body of healthy adults (airway, skin, oral cavity, gastrointestinal tract, and vagina). Several 2 specific body sites were sampled within each major region (18 in total), and as the volunteers were clinically verified to be free of overt disease in all body regions, this study is known as the healthy adult cohort study. Three hundred adult volunteers were enrolled at two US clinical centers. These included equal numbers of 18-40 year old men and women, 20% of whom self-identified as a racial minority, and 11% selfidentified as Hispanic. Exclusion and inclusion criteria, clinical sampling procedures, and the corresponding clinical metadata, can be found at the NCBI database of Genotypes and Phenotypes (dbGaP, http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000228). Of the 300 volunteers in this study, over 2/3 were sampled twice and 1/3 were sampled a third time, over approximately two years. Among the 18 total body sites sampled, the oral cavity had the largest number of sites (9) and all body sites were directly sampled except for the gastrointestinal tract, for which stool served as a proxy. Blood was collected for serum and for bulk DNA for human subject whole genome sequencing, and lymphocytes were harvested for the preparation of cell lines; these specimens are being held for future analysis and distribution when the larger research community expresses interest in these resources. Of the over 10,000 primary microbiome specimens collected for the full cohort, all have been sequenced for the 16S rRNA gene taxonomic marker. Metagenomic sequence data has additionally been generated from approximately one thousand of the nucleic acid samples. A portion of the full set of 16S data and of the metagenomic data was targeted by the HMP research consortium for a global analysis. In 2012, these analyses were published in Nature (The Human Microbiome Project Consortium, 2012a, 2012b) and in the HMP collection of related papers in PLoS (http://www.ploscollections.org/article/browseIssue.action?issue=info:doi/10.1371/issue.pcol.v01.i13#c over). As of this writing, over 300 publications from analysis of the healthy cohort study data and from other studies in the program are in PubMed and cite HMP support. A current list can be found at https://commonfund.nih.gov/hmp/publications.aspx. HMP Demonstration Projects A third key resource from this program are the HMP Demonstration Projects, designed to evaluate microbiome characteristics in health conditions or disease states with putative microbiome associations. Many complex diseases appear to have a microbiome component, and these projects were designed to characterize the microbiome in such cases in order to develop a reference dataset of microbiome properties associated with specific disease and clinical phenotypes. Eleven Demonstration Project studies were supported in the program, including six projects on microbiome-associated gastrointestinal diseases (Crohn’s disease, ulcerative colitis, pediatric inflammatory bowel syndrome, neonatal necroticizing enterocolitis, and esophageal adenocarcinoma), three on urogenital status (i.e., microbiome characteristics associated with bacterial vaginosis, reproductive history, sexual history and circumcision), and two on microbiome-associated skin diseases (atopic dermatitis and psoriasis). Almost all of the studies include 16S rRNA gene and metagenomic sequencing, and some also include functional data from the microbiome such as microbial community gene expression, proteomics, or metabolomics. The Demonstration Projects sequence datasets and study descriptions can be found at NCBI Bioprojects (http://www.ncbi.nlm.nih.gov/bioproject/46305). HMP Phase Two It was recognized during the analysis of the healthy cohort metagenomic data that there appeared to be millions of unique microbial genes in the human microbiome and this brought into question the genetic 3 potential of these microbial communities. What products and pathways do these microbial genes encode for? It was not feasible to decode all of the transcripts, gene products and metabolites of the global human microbiome. However, a more limited effort to collect biological properties of the microbiome in well-characterized cohorts of putative microbiome-associated diseases or health conditions became the focus of the second phase of the HMP program. The data from these studies are intended to serve as model systems for microbiome-associated disease or health conditions and so of broad interest to the larger research community. The properties of particular interest include the microbial community gene expression profiles (metatranscriptome), the proteins (metaproteome) and the metabolic products of microbial activity (metabolome). These data will be combined to create an integrated dataset of microbiome and host properties of subjects as a community resource. This dataset will provide the research community the opportunity to evaluate which microbiome properties or combination of properties are the most informative when investigating the role of the microbiome in disease or in health. Future Directions for Human Microbiome Research Though the HMP may be the most visible example of NIH interest in the human microbiome, there are currently 16 NIH Institutes and Centers which have microbiome-related research programs or are developing new microbiome programs. A recent three-day meeting brought together the NIH and the larger research community to discuss the state of the science for this field and to identify the gaps, needs and challenges for advancing this field over the next ten years. The videotaped talks and slides are available here http://www.genome.gov/27554404. One of the crucial needs called out by these meeting participants was the desire for large, longterm cohort studies which could serve as a platform from which numerous investigations could address microbiome development, variability of the microbiome across populations, temporal changes, and functional properties in response to diet or disease or other perturbations or interventions. Genome sequences of the cohort subjects would provide invaluable data for integration with the microbiome properties. Opportunities for cohort studies may soon become available; for example, a federal collaboration between the NIH, the CDC and the EPA is currently developing the National Children’s Study (http://www.nationalchildrensstudy.gov), which plans to follow 100,000 children from birth to 21 years of age. The pilot phase of the NCS is exploring the inclusion of microbiome samples during this large study. These and other cohort studies could provide the ideal framework from which to analyze the microbiome from birth throughout lifetimes in diverse populations. References Costello, E.K., Lauber, C.L., Hamady, M., Fierer, N., Gordon, J.I., and Knight, R. (2009). Bacterial community variation in human body habitats across space and time. Science 326, 1694-1697. Hooper, L.V. and J. I. Gordon. (2001). Commensal host-bacterial relationships in the gut. Science 292, 1115-1118. The Human Microbiome Project Consortium (2012a). Structure, function and diversity of the healthy human microbiome. Nature 486, 207-214. The Human Microbiome Project Consortium (2012a).A framework for human microbiome research. Nature 486, 215-219. 4 Koenig, J.E., Spor, A., Scalfone, N., Fricker, A.D., Stombaugh, J., Knight, R., Angenent, L.T., and Ley, R.E. (2011). Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 108 Suppl 1, 4578-4585. Nelson, K.E., Weinstock, G.M., Highlander, S.K., Worley, K.C., Creasy, H.H., Wortman, J.R., Rusch, D.B., Mitreva, M., Sodergren, E., Chinwalla, A.T., et al. (2010). A catalog of reference genomes from the human microbiome. Science 328, 994-999. Round, J.L., and Mazmanian, S.K. (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9, 313-323. 5