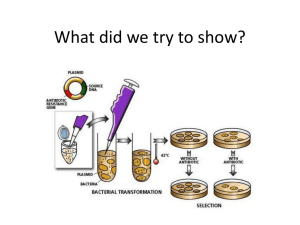
Activity 4.1.2 Protein Factories Introduction
advertisement

Activity 4.1.2 Protein Factories Introduction For years, Diana Jones relied on insulin shots to get her through the day. Countless needle sticks measured her sugar levels and alerted her to the need for more medication. Even though life is a bit simpler now that she has her insulin pump, Mrs. Jones’s survival is still directly linked to the medicine being pumped into her bloodstream. What she does not realize, however, is that the protein that is keeping her body regulated can be produced by tiny living organisms, particularly bacteria. When most people hear the word bacteria, they probably think about getting sick or about destroying these invaders with antibiotics. However, bacteria are a very helpful tool in genetic engineering and in the production of medication. These living cells replicate quickly and their unique structure provides scientists with the ability to “highjack” their molecular machinery and insert a new gene – the code for a new protein. Genetically engineered bacteria can be used as tiny living pharmaceutical factories and can produce many medications and drugs that help maintain health and homeostasis in the human body. Remember that a gene is a piece of DNA that provides the instructions for making a protein. Genes can be cut out of human, animal or plant DNA and placed into bacteria in small rings of DNA called plasmids. Bacteria in nature can pass plasmids back and forth to share beneficial genes. In Lesson 1.2 you learned that this swap of genetic material can lead to shared traits such as antibiotic resistance. In the lab, scientists can alter plasmids to include the code for a protein of interest and insert this ring of DNA into bacterial cells. The result of this genetic engineering is that the recipient organism now has the instructions to make a new protein. In 1978, scientists from the California based company Genentech succeeded in manipulating bacterial DNA to produce human insulin. The final product was released to the public in the early 1980s, bringing relief to many diabetics. As you recall from Unit 1, scientists can use the tools of molecular biology to “cut and paste” genes of interest into bacterial plasmids. These plasmids can then be inserted into bacteria through the process of bacterial transformation. Once the new gene is inside the cell, the bacteria multiplies and the new bacterial clones crank out many copies of a desired protein product, such as insulin. In this experiment, you will not be using bacterial cells to produce insulin. Instead, you will use bacterial transformation to insert the genetic information necessary to produce another protein, green fluorescent protein (GFP), into the cells. Since this newly produced protein glows a bright green, it is easy to gauge the success of the experiment and really see the process of transformation at work. Equipment Bio-Rad pGLO Bacterial Transformation Kit o E. coli bacteria o pGLO™ plasmid o LB bacterial growth plates o LB bacterial growth plates with ampicillin o LB bacterial growth plates with ampicillin and arabinose o Disposable sterile pipets o Microcentrifuge tubes o Foam racks © 2010 Project Lead The Way, Inc. Medical Interventions Activity 4.1.2 Protein Factories – Page 1 o CaCl2 transformation solution Computer with Internet access Laboratory journal Transformation Kit – Quick Guide handout Transformation Efficiency worksheet Handheld UV light or transilluminator Microcentrifuge tube rack o Inoculation loops Water bath – 42°C Incubator – 37°C Permanent black marker Ice and ice container Colored pencils or markers Sterile water (optional) Procedure Part I: Bacterial Transformation 1. Turn to the diagram of a bacterial cell you drew and labeled in your laboratory journal in Unit 1. Review the function of key structures in this cell and locate a plasmid. 2. Define the term plasmid in your laboratory journal. Write a list of at least three reasons why plasmids and bacterial cells are a helpful tool in genetic engineering. Use the Internet or your notes from Unit 1 if you need additional information. 3. View the animations located at the Explore More – Iowa Public Television site http://www.iptv.org/exploremore/ge/what/insulin.cfm to learn how recombinant DNA techniques and plasmids can be used to produce human insulin. Outline the steps of the process in your laboratory journal. 4. Answer Conclusion questions 1-3. 5. Brainstorm how you could use recombinant DNA technology to make normal E. coli bacterial cells glow green. Where in nature could you find a source for a glowing protein? Discuss your ideas with a partner. 6. Note that in this laboratory experiment, the plasmid containing the gene of interest, GFP, has already been produced for you using recombinant DNA techniques. You will complete bacterial transformation to move this plasmid into bacterial cells. With the new gene, bacterial cells can now produce the green fluorescent protein. 7. Visit the Dolan DNA Learning Center DNA Interactive site found at http://www.dnai.org/b/index.html to review the process of transformation. 8. Click on the Techniques tab at the bottom of the screen and then choose Transferring and Sorting. 9. View the Transformation animation. Take notes in your laboratory journal as you view the animation. 10. Summarize the goal of the experiment in a well-crafted paragraph in your laboratory journal. Your summary should include the terms listed below. Underline each term. o o o o o o Aequorea Victoria jellyfish gene pGLO plasmid green fluorescent protein o o o o o o ampicillin arabinose E. coli bacteria ultraviolet light chemical transformation heat shock © 2010 Project Lead The Way, Inc. Medical Interventions Activity 4.1.2 Protein Factories – Page 2 11. Draw the pGLO plasmid map, the diagram showing the location of important genes, in your laboratory journal. Label each of the three key genes in a different color. Provide a one sentence description of the function of each gene. Make sure to mention how the gene relates to the protein that is being produced. 12. Answer Conclusion questions 4-5. 13. Obtain a Transformation Kit – Quick Guide handout and the following plates from your teacher: o (1) LB bacterial growth plate o (2) LB bacterial growth plates with ampicillin o (1) LB bacterial growth plate with ampicillin and arabinose 14. Follow the directions on the Quick Guide reference sheet to complete the experiment. Use the drawings located on the right side of the page as a visual reference for each step. Make sure to clearly label each plate. Note whether the media contains ampicillin or arabinose (or both) and whether you are plating out bacteria that contain the pGLO plasmid or that do not contain the plasmid. Depending on the length of your lab period, your teacher may ask you to stop at Step 9. The sample tubes can be incubated overnight at room temperature and the experiment resumed on the following day. 15. At the conclusion of the experiment, stack your labeled plates and place the stack inverted in the 37°C incubator. Allow bacteria to incubate for approximately 24 hours. 16. Predict which of the four plates should show growth. Predict which of the four plates should glow green when exposed to UV light. Write your predictions down in your laboratory journal and explain your reasoning. 17. Remove the stack of plates from the incubator after 24 hours. 18. Copy the data table shown below into your laboratory journal and follow Steps 21 - 23 to make observations and to fill in laboratory data. Growth? (Y/N) If yes, include a colony count. Glowing? (Y/N) Other Observations -pGLO LB -pGLO LB/amp +pGLO LB/amp +pGLO LB/amp/ara 19. Observe the bacterial growth on each plate. Carefully count the number of colonies that are visible. You may want to use a black marker to dot each colony you count. This technique may help you track the colonies you have already counted. 20. Place each plate on the UV transilluminator or use a handheld UV light to determine if the colonies do or do not glow green. 21. Record any other relevant observations in your data table. 22. Compare the results to the predictions you documented in your laboratory journal. Discuss your results with the class. 23. Answer Conclusion questions 6-9. © 2010 Project Lead The Way, Inc. Medical Interventions Activity 4.1.2 Protein Factories – Page 3 Part II: Calculating Transformation Efficiency The first genetically engineered human insulin was produced in the early 1980s. This technology was a huge achievement in diabetes therapy as it allowed for the production of nearly unlimited quantities of human insulin. In order for the process to work efficiently, it is necessary to transform as many cells as possible in each round of bacterial transformation. The more cells that are successfully transformed to produce the desired protein, the more patients can be helped with the therapy. Transformation efficiency is a quantitative measurement that gives scientists an indication of how successful they were in getting the desired DNA into the bacterial cells. Transformation efficiency in this experiment represents the total number of bacterial cells that express the new protein (in your case, GFP), divided by the total amount of DNA used in the experiment. This value tells scientists the number of bacterial cells transformed by one microgram of DNA and is calculated by the general formula: Transformation efficiency = Total number of cells growing on the plate Amount of DNA spread on the plate (in µg) Before you can calculate the efficiency of your experiment, you will need two pieces of information: the total number of glowing colonies on your LB/amp/ara plate and the total amount of pGLO plasmid DNA spread on the LB/amp/ara plate. You counted the number of colonies in part I of the experiment. Follow the directions below to determine the amount of DNA spread and to calculate the overall transformation efficiency. 24. Obtain a Transformation Efficiency worksheet from your teacher. 25. Follow the steps on the worksheet to complete the calculations. Use procedural information found in the Quick Guide as well as the data table from Part I of the laboratory experiment to assist you with your calculations. 26. Write the formula for transformation efficiency in your laboratory journal. 27. Clearly show your final calculations for transformation efficiency in your laboratory journal. Report your final value using scientific notation. 28. Note that transformation efficiency values for the experiment completed in Part I are generally between 8.0 x 102 and 7.0 x 103 transformants per microgram of DNA. Compare the transformation efficiency of your experiment to this value and describe your findings in your lab journal. 29. Compare your transformation efficiency to that of another lab group. 30. Answer Conclusion questions 10 and 11. Part III: GFP as a Medical Intervention 31. View the Virginia Commonwealth University Secrets of the Sequence video A Green Light for Biology – Making the Invisible Visible at http://www.pubinfo.vcu.edu/secretsofthesequence/playlist_frame.asp to explore how the protein GFP can be used to “tag” proteins of interest. 32. Read the article Lighting Up the Lab found at the UCSC Science Communication program site at http://sciencenotes.ucsc.edu/0001/glow.htm. © 2010 Project Lead The Way, Inc. Medical Interventions Activity 4.1.2 Protein Factories – Page 4 33. Use the information from the video and the article to list and describe at least three useful applications of GFP in cellular biology and medical science in your laboratory journal. Think about how GFP qualifies as a medical intervention. 34. Answer the remaining Conclusion question. Conclusion 1. Why are bacteria a good choice for producing many copies of a particular gene in a short period of time? 2. How are restriction enzymes and ligase utilized in recombinant DNA technology? 3. Explain why it is necessary to use the same restriction enzyme to cut the desired gene from the source and to cut the plasmid. Include a drawing or diagram in your answer. 4. Explain the role of arabinose in the bacterial transformation experiment. 5. How do the heat shock and the calcium chloride assist plasmid insertion in this chemical transformation? Relate your answer to the charge on DNA and to the charge on the plasma membrane of a cell. 6. How do the results of this experiment illustrate the relationship between DNA, proteins and a trait? 7. How would a genetic engineer distinguish between the bacteria containing the new plasmid DNA and those bacteria lacking this plasmid DNA? 8. Very often an organism’s traits are influenced by a combination of its genes and its environment. What two factors must be present in the bacteria’s environment for you to see the color green? Explain your answer. 9. What advantage would there be for an organism to be able to turn on or off particular genes in response to certain situations? 10. Provide at least two explanations for why transformation efficiency varied from group to group. Think about sources of error in the experiment. What could you do to maximize transformation efficiency? 11. Explain how a value such as transformation efficiency might relate to the business side of using bacteria to produce proteins. 12. The bacterial cells produce many more proteins than just the GFP. How do you think we can separate the GFP from the other proteins in the cell? © 2010 Project Lead The Way, Inc. Medical Interventions Activity 4.1.2 Protein Factories – Page 5