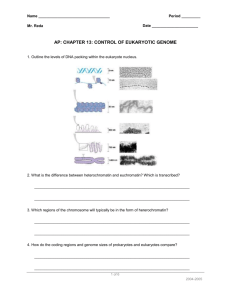
Stickleback Evolution Research Video Notes: Learning goals: After
advertisement

Stickleback Evolution Research Video Notes: Learning goals: 1. After watching a presentation of research, determine background information, hypothesis, and basic techniques. 2. After watching a presentation of research, be able to describe the main experimental results and explain how this supports the hypothesis. The answers to the following questions can be found in the video. Nearly identical questions will be asked in the online basics quiz. 1. In the first 5 minutes of the video the background of the is established. a. What change in climate allowed sticklebacks to populate new areas? b. What happened to populations in these new areas when geology pushed the land upward? c. What phenotypic change occurred in the fish? About how many years have elapsed? d. What environmental pressures (predators) likely caused this change? e. “Changes in form are ultimately due to changes in ____________ “ 2. The two main scientists shown in the video are Dr. Michael Bell and Dr. David Kingsley. What strengths do each bring to the research? Michael Bell: http://life.bio.sunysb.edu/ee/belllab/research.html David Kingsley: http://kingsley.stanford.edu/sticklebacks.html Stickleback Evolution Page 1 The first experiment is described at about minute 5. The video attempts to describe the process of genome-wide linkage mapping. Let’s look at some take-home messages: 3. When a pelvic-complete parent is crossed with a pelvic-reduced parent, all the offspring have complete pelvises. a. What are the two alleles? Which is dominant? b. What is the genotype of the F1 generation? In linkage mapping, DNA markers (rare sequences of DNA bases) are used that have a known location on a known chromosome. Usually several dozen markers are used. When a hybrid fish generates eggs or sperm, crossing over occurs. An adult fish can be examined to see if it has a complete pelvis, and its genome analzyed for each marker. If a chromosome marker is ALWAYS seen with a phenotypic trait, even after several different instances of crossing over, then the gene associated with the phenotype is likely found close to that marker. 4. The pelvic-reduced phenotype was found to co-occur with a marker on chromosome 7 very near the developmental gene called: ________________ Researchers then used gene sequencing to compare the coding region of the gene in fish with different phenotypes. 5. Was there a mutation in the coding sequence of this gene in the pelvic-reduced fish? 6. At Minute 7, Kingsley’s team moves on to see if there are changes in gene regulation rather than gene coding. The blue dye binds to what molecule in the fish embryo? Circle one. Pitx1 DNA Pitx1 mRNA Pitx1 protein 7. Where did expression of the gene remain in freshwater fish? Where was it turned off? 8. What was Kingsley’s hypothesis for this result? Stickleback Evolution Page 2 To test the hypothesis, they needed to find a regulatory gene that is only active in pelvic-complete fish. they took fragments of DNA near the Pitx1 gene from a pelvic-complete fish and attached a new DNA sequences to each that codes for a protein that fluoresces green. They re-injected the fragments into fish embryos, keeping track of which DNA fragments went into which fish. Eventually, a fish making its pelvic girdle turned on its own Pitx1 enhancer, PLUS the specially-labeled Pitx1 enhancer on the DNA fragment. 9. A fish with a fluorescent green pelvis is expressing which? Circle one. Pitx1 regulatory gene Pitx1 coding gene 10. What did they find when they compared this region from pelvic-complete fish to pelvic-reduced fish? Was there a mutation? What kind? 11. In the last experiment (time 10:09), they took this same fragment of DNA and injected it into an embryo with pelvic-reduced parents. What was the result? Why? 12. From about Minute 11 to the end of the video, the researchers describe the implications of this project, and what new things they are learning. Write down an interesting fact from this part of the video to share in the online quiz. Be sure to bring these completed notes to class. There is also an optional online activity you may want to try at the end of the in-class activity. If that sounds intriguing, bring a laptop or smartphone/tablet. Stickleback Evolution Page 3 Class Notes: Learning goals: 1. Verify understanding of research projects by discussing with group members 2. Use new understanding to interpret research data and predict results. 1. Compare your answers on the video notes to those of your group members. Students who do not bring their completed notes cannot participate in this activity. 2. The following experiment was from the same paper as used in the video. Let’s explore a new experiment more closely. Geneticists use two terms to talk about gene regulation: cis-regulatory elements and trans-acting factors. Cis- regulatory elements Trans-acting factors “cis” means same, as in “same “trans” means other, as in “from chromosome” as the target gene another molecule” Made of DNA Can refer to transcription factors, or Usually within 200 base pairs of the the genes that code for them. promoter of the transcription start Separate from the target gene site of the target gene The following diagram is meant to represent the activity of the Pitx1 gene in a stickleback embryo: Are activators considered cis or trans? _____________________ Are switches considered cis or trans? _____________________ Stickleback Evolution Page 4 The paper says this: Although Pitx1 represents a strong candidate gene for pelvic reduction, other genes in the larger chromosome region could be the real cause of pelvic loss, leading to secondary or trans-acting reduction of Pitx1 expression. What does this mean in introductory biology English? Convert this to a statement that indicates an activator may be the cause: The paper then goes on to say: To test this possibility, we generated F1 hybrids between pelvic-complete (Friant Low (FRIL) and pelvic-reduced (Paxton Lake Benthic (PAXB)) sticklebacks. F1 hybrid fish develop pelvic structures and contain both Pitx1 alleles in an identical trans-acting environment. Here is part of the figure from the paper, and the figure legend: Alleles of Pitx1 from pelvic-complete (FRIL and LITC) and pelvicreduced populations (PAXB) were combined in F1 hybrids, and brain and pelvic tissues were isolated so as to compare the expression of either the LITC or PAXB allele normalized to the level of expression of the FRIL allele in the same trans-acting environment. Cross 1: LITC x FRIL This cross is the control, which shows the effect of “being crossed with a FRIL.” The main measurement is how much Pitx1 mRNA is produced by the complete-pelvis or reduced-pelvis allele. How different/similar are the Pitx1 coding regions of all three alleles? Stickleback Evolution Page 5 Cross 2: PAXB x FRIL Do F1 embryos contain the activators for both head and pelvis Pitx1? How do you know? How much Pitx1 would you expect this embryo to make in head tissue? Reduced Normal If PAXB normally have reduced Pitx1 expression because of a loss of a functional activator, how much Pitx1 will be made in these F1 embryos? Reduced Normal How much PAXB Pitx1 mRNA would you expect to see if instead their lack of pelvic spines were caused by a mutation in the pelvis switch? Reduced Normal To the right is the experimental result. Restate in your own words: a. The hypothesis being tested, and b. What these results demonstrate This video comes with several different resources on the website Bioactive.org. They include a virtual lab that allows you to try out some of the research techniques. If you finish early, use a laptop or smartphone to try it out: http://media.hhmi.org/biointeractive/vlabs/stickleback/index.html or http://goo.gl/EbzU0V Reference: Adaptive Evolution of Pelvic Reduction in Sticklebacks by Recurrent Deletion of a Pitx1 Enhancer. Yingguang Frank Chan, Melissa E. Marks, Felicity C. Jones, Guadalupe Villarreal, Jr., Michael D. Shapiro, Shannon D. Brady, Audrey M. Southwick, Devin M. Absher, Jane Grimwood, Jeremy Schmutz, Richard M. Myers, Dmitri Petrov, Bjarni Jónsson, Dolph Schluter, Michael A. Bell, and David M. Kingsley. Science 15 January 2010: 327 (5963), 302-305. [DOI:10.1126/science.1182213] Stickleback Evolution Page 6