Scalability of DryadLINQ
advertisement
Scalability of DryadLINQ
Haomin Xiang
Student
2001 E.lingelbach bane
Apt.# 440 Bloomington, IN
8123609957 47408
xhmsun1987@gmail.com
Abstract
DryadLINQ is a system, which enables a brand new
programming model for large scale distributed
computing by providing a set of language extensions.
This paper describes the scalability of
implementation of the DryadLINQ and it evaluates
DryadLINQ on several programs implemented by
DryadLINQ, that is, CAP3 DNA sequence assembly
program [1], High Energy Physics data analysis,
CloudBurst [2] - a parallel seed-and- extend readmapping application and compare their scalability
with the same applications implemented by Hadoop.
Categories
Descriptors
and
Subject
Describe
the
system
architecture
of
DryadLINQ, and show the scalability of
DryadLINQ by comparing program implemented
by DryadLINQ with those implemented by
Hadoop and CGL-MapReduce.
General Terms
Performance, Language
Keywords
2. DryadLINQ and Hadoop
The DryadLINQ system is designed to materialize the
idea that a wide array of developers could compute on
large amounts of data effectively and suffer little
performance loss in scaling up with modest effort.
2.1 DryadLINQ System Architecture
LINQ programs are complied into distributed
computations by DrtadLINQ, and it would then
running on the Dryad cluster-computing infrastructure.
A Dryad job is a directed acyclic graph where each
edge represent a data channel and vertexes are
programs.
The execution of a Dryad job is maneuvered by a
centralized “job manager.” The job manager is
responsible for:
First, Materializing a job’s dataflow graph;
Second, Scheduling processes on cluster computers;
Third, Enabling fault-tolerance by re-executing slow
or failed processes;
Forth, Monitoring the job and collecting dates; and
Sixth, Dynamically transforming the job graph
according to the rules defined by users.
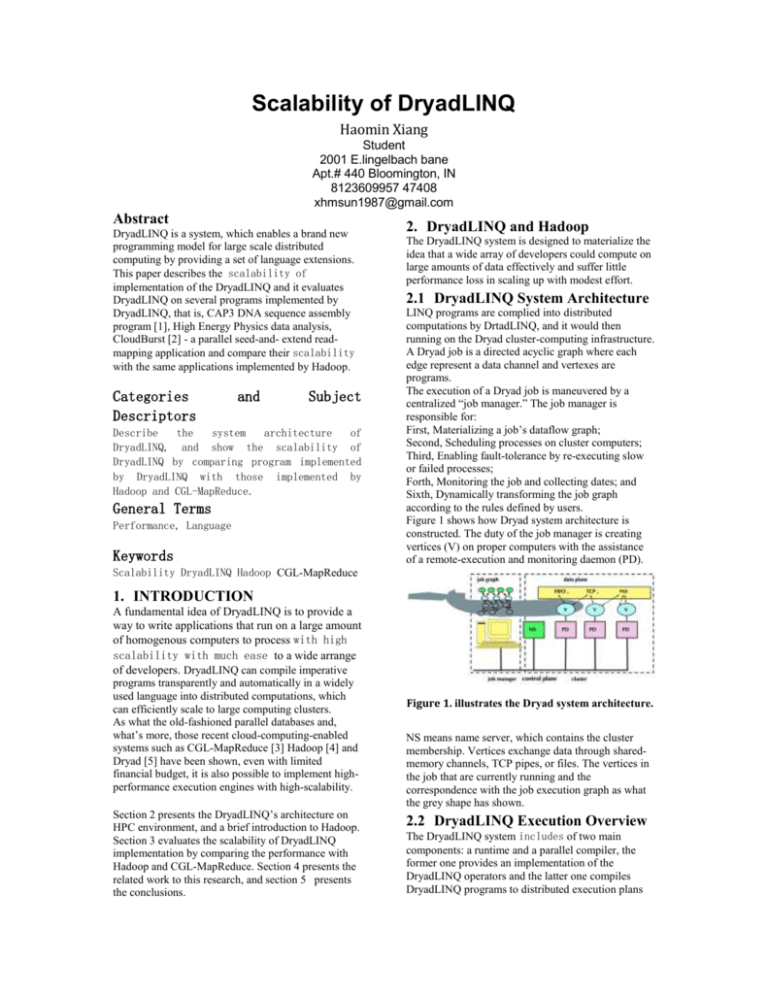
Figure 1 shows how Dryad system architecture is
constructed. The duty of the job manager is creating
vertices (V) on proper computers with the assistance
of a remote-execution and monitoring daemon (PD).
Scalability DryadLINQ Hadoop CGL-MapReduce
1. INTRODUCTION
A fundamental idea of DryadLINQ is to provide a
way to write applications that run on a large amount
of homogenous computers to process with high
scalability with much ease to a wide arrange
of developers. DryadLINQ can compile imperative
programs transparently and automatically in a widely
used language into distributed computations, which
can efficiently scale to large computing clusters.
As what the old-fashioned parallel databases and,
what’s more, those recent cloud-computing-enabled
systems such as CGL-MapReduce [3] Hadoop [4] and
Dryad [5] have been shown, even with limited
financial budget, it is also possible to implement highperformance execution engines with high-scalability.
Section 2 presents the DryadLINQ’s architecture on
HPC environment, and a brief introduction to Hadoop.
Section 3 evaluates the scalability of DryadLINQ
implementation by comparing the performance with
Hadoop and CGL-MapReduce. Section 4 presents the
related work to this research, and section 5 presents
the conclusions.
Figure 1. illustrates the Dryad system architecture.
NS means name server, which contains the cluster
membership. Vertices exchange data through sharedmemory channels, TCP pipes, or files. The vertices in
the job that are currently running and the
correspondence with the job execution graph as what
the grey shape has shown.
2.2 DryadLINQ Execution Overview
The DryadLINQ system includes of two main
components: a runtime and a parallel compiler, the
former one provides an implementation of the
DryadLINQ operators and the latter one compiles
DryadLINQ programs to distributed execution plans
In Steps 1–2, executing a .NET user application. As
LINQ deferred execution, expressions are
accumulated in a DryadLINQ expression object, the
application invoking a method that materializes the
output dataset triggering their actual execution. After
the triggering, DryadLINQ
allows the data to be dispersed to all the nodes. In
order to improve the overall I/O bandwidth,
Hadoop takes the data locality into major concern
when schedules the MapReduce computation tasks.
The outputs of the map tasks would be accessed by
the reduce tasks via HTTP connections, before this,
Figure 2. shows the flow of execution when a
program is executed by DryadLINQ.
takes over and compiles the LINQ expression into a
distributed execution plan which can be
understood by Dryad (Step 3). This step performs
the majority of the compiling work, containing:
(a) Decomposing the expression into sub expressions,
each to be run in a separate Dryad vertex;
(b) Generating code and static data for the remote
Dryad vertices; and
(c) Generating serialization code for the required data
types.
At Step 4, a custom, DryadLINQ-specific, Dryad job
manager is invoked.
Dryad takes control at Step 5. By using the plan
created in Step 3, it creates the job graph, and
schedules and creates the vertices when cluster
resources are available. Each Dryad vertex runs a
program, which specific for it created in Step 3 by
DryadLINQ. After the Dryad job completes
successfully, it returns the control back to DryadLINQ
at Step 8 and writes the data to the output table(s).
2.3 Hadoop and HDFS
When comparing to Google’s MapReduce runtime,
Apache Hadoop has a similar architecture to it,
where Apache Hadoop accesses data via HDFS,
which maps all the local disks of the compute nodes
to a single file system hierarchy, and the file system
Figure 3. Comparison of features supported by
Dryad and Hadoop of heterogeneous compute
nodes.
they would be stored in local disks.
Although this approach enables the fault-tolerance
mechanism in Hadoop, however, it imposes
considerable communication burden to the
intermediate data transformation, especially for the
cases that executing the applications, which
produce small intermediate results frequently.
3. Experimental Evaluations
3.1 Hardware Configuration
Table 1. Hardware Configuration
Feature
Linux
Cluster
Window
Cluster
CPU
#CPU
#Cores
Memory
#disk
Network
Operating
System
#Nodes
Intel(R )
Xeon (R )
CPU L5420
2.50GHz
2
8
32GB
1
Giga bit
Ethernet
Red Hat
Enterprise
Linux
Server -64
bit
32
Intel(R )
Xeon (R )
CPU L5420
2.50GHz
2
8
16GB
2
Giga bit
Ethernet
Windows
Server
Enterprise
-64bit
scheduling of individual CAP3 executable in a
given node is not optimal.
The reason why the utilization of CPU cores is not
optimal is that when an application is scheduled,
DryadLINQ uses the number of data partitions as a
guideline to schedules the number of vertices to to
the nodes rather than individual CPU cores under
the assumption that the underlying PLINQ
runtime would handle the further parallelism
available at each vertex and utilize all the CPU
cores by chunking the input data.
However, our input for DraydLINQ is only the
names of the original data files, it has no way to
determine how much time the Cap3.exe take to
process a file, and hence the chunking of records at
32
3.2 CAP3
CAP3 is a DNA sequence assembly program that
performs several major assembly steps to a given set
of gene sequences.
Cap3.exe (Input) output + Other output files
As what have been shown above, the program reads
gene sequences, each of which needs to be processed
by the CAP3 program separately, from an input file
and writes its output to several output files and to the
standard output.
DryadLINQ application only needs to know the input
file names and their locations for it executes the CAP3
executable as an external program.
(1) Each node of the cluster stores roughly the same
number of input data files by dividing the input data
files among them;
(2) Each node creates a data partition which
containing the names of the original data files
available in that node;
(3) Individual data-partitions stored in each node
are pointed to the late-created Dryad partitionedfile.
After what has been shown above, a DryadLINQ
program would be created to read the data file
names from the provided partitioned-file, and
execute the CAP3 program.
However, a suboptimal CPU core utilization, which
is highly unlikely for CAP3, is noticed.
A trace of job scheduling in the HPC cluster
revealed that the utilization of CPU cores of the
Figure 4. Performance of different
implementations of CAP3 application.
PLINQ would not lead to optimizing of the
schedule of tasks.
Figure 4 and 5 show comparisons of performance
and the scalability of all three runtimes for the
CAP3 application.
DryadLINQ does not schedule multiple concurrent
vertices to a given node, but one vertex at a time.
Therefore, a vertex, which uses PLINQ to schedule
some non- homogeneous parallel tasks, would have a
running time equal to the task, which takes the longest
time to complete.
In contrast, we can set the maximum and minimum
number of map and reduce tasks to execute
concurrently on a given node in Hadoop, so that it will
utilize all the CPU cores.
The performance and the scalability graphs indicate
that the DryadLINQ application, the Hadoop and
CGL-MapReduce versions of the CAP3 application
work almost equally well for the CAP3 program.
produce the final histogram of identified features.
The results of this analysis among the performance
of three runtime implantations are shown in
Figure 6.
The results in Figure 6 implicates that
compared to DraydLINQ and CGL-MapReduce
implementations, Hadoop implementation has a
remarkable overhead which is mainly due to
differences in the storage mechanisms used in these
frameworks.
Figure 5. Scalability of different
implementations of CAP3
3.3 HEP
HEP is short for High-energy physics.
In HEP application, the input is available as a
collection of large number of binary files, which will
not be directly accessed by the DryadLINQ program,
so we have to
(1) Give each compute node of the cluster a
division of input data manually, and
(2) Put a data-partitions, which stores only the
file names, in a given node.
The first step of the analysis requires applying a
function coded in ROOT script to all the input files:
[Homomorphic]
ApplyROOT(string filename){..}
IQueryable<HistoFile>histograms =
DataFileName. Apply(s =>
ApplyROOT(s));
the Apply operation allows a function to be applied to
an entire data set, and produce multiple output
values ,so in each vertex the program can access a data
partition available in that node.
Inside the ApplyROOT() method, the program
iterates over the data set and groups the input data files,
and execute the ROOT script passing these files names
along with other necessary parameters, what’s more, it
also saves the output, that is, a binary file containing a
histogram of identified features of the input data, in a
predefined shared directory and produces its location
as the return value.
In the next step of the program, we perform a
combining operation to these partial histograms by
using a homomorphic Apply operation and to
those collections of histograms in a given data
partition by using another ROOT script, finally to the
output partial histograms produced by the previous
step by the main program. The last combination would
Figure 6. Performance of different
implementations of HEP data analysis
applications
HDFS can only be accessed using C++ or Java clients,
and the ROOT data analysis framework is not capable
of accessing the input from HDFS.
In contrast, both Dryad and CGL-MapReduce
implementations’ performance are improved
significantly by the ability of reading input from the
local disks.
What’s more, in the DryadLINQ implementation, the
intermediate partial histograms are stored in a shared
directory and are combined during the second phase as
a separate analysis. In CGL-MapReduce
implementation, the partial histograms are directly
transferred to the reducers where they are saved in
local file systems and combined.
This difference can explain the performance difference
between the CGL-MapReduce implementation and the
DryadLINQ implementation.
3.4 CloudBurst
CloudBurst is a Hadoop application that performs a
parallel seed-and-extend read-mapping algorithm to
the human genome and other reference genomes.
CloudBurst parallelizes execution by seed, that is
the reference and query sequences would be
grouped together and sent to a reducer for further
analysis if they sharing the same seed.
CloudBurst is composed of a two-stage MapReduce
workflow:
(1) Compute the alignments for each read with at
most k differences where k is user specified.
(2) Report only the best unambiguous alignment for
each read rather than the full catalog of all
alignments.
An important characteristic of the application is that
the variable amount of time it spent in the reduction
phase. This characteristic can be a limiting factor to
scale, depending on the scheduling policies of the
framework running the algorithm.
In DryadLINQ, the same workflow is expressed as
follows:
MapGroupByReduceGroupByReduce
DryadLINQ runs the whole computation as a whole
rather than two separate steps followed by one
another.
The reduce function would produces one or more
alignments after receives a set of reference and
query seeds sharing the same key as input. For each
input record, query seeds are grouped in batches,
and in order to reduce the memory limitations each
batch is sent to an alignment function sequentially.
We developed another DryadLINQ implementation
that can process each batch in parallel assigning
them as separate threads running at the same time
using .NET Parallel Extensions.
The results in Figure 7s show that all three
implementations follow a similar pattern although
DryadLINQ is not fast enough especially when nodes
number is small.
Figure 7. Scalability of CloudBurst with
different implementations
The major difference between DryadLINQ and
Hadoop implementations is that in DraydLINQ, even
though PLINQ assign records to separate threads
running concurrently, the cores were not utilized
completely; conversely, in Hadoop each node starts
reduce tasks which has number equal to one’s cores
and each task ran independently by doing a fairly
equal amount of work.
Another difference between DryadLINQ and Hadoop
implementations is the number of partitions created
before the reduce step. Since Hadoop creates more
partitions, it balances the workload among reducers
more equally.
If the PLINQ scheduler worked as expected, it would
keep the cores busy and thus yield a similar load
balance to Hadoop.
In order to achieve this, one can try starting the
computation with more partitions aiming to schedule
multiple vertices per node. However, DryadLINQ runs
the tasks in order, so it would wait for one vertex to
finish before scheduling the second vertex, but the first
vertex may be busy with only one record, and thus
holding the rest of the cores idle.
So the final effort to reduce this gap would be the
using of the .NET parallel extensions to fully utilizing
the idle cores, although it is not identical to Hadoop’s
level of parallelism.
Figure 6 shows the performance comparison of three
runtimes implementation with increasing data size.
Both implementations of DryadLINQ and Hadoop
scale linearly, and the time gap is mainly related to
what has been explained above: the current limitations
with PLINQ and DryadLINQ’s job scheduling polices.
Figure 6. Performance comparison of DryadLINQ
and Hadoop for CloudBurst.
Relate work
2000s sees a large amount of activity in architectures
for processing large-scale datasets. One of the earliest
commercial generic platforms for distributed
computation was the Teoma Neptune platform [6],
which introduced a map-reduce computation paradigm
inspired by MPI’s Reduce operator. The Hadoop
open-source port of MapReduce slightly extended the
computation model, separated the execution layer
from storage, and virtualized the execution. The
Google MapReduce [7] uses the same architecture.
NetSolve [8] provides a grid-based architecture for a
generic execution layer. DryadLINQ has a richer set of
operators and better language support than any of
those above.
At the storage layer a variety of very large-scale
simple databases have appeared, including Amazon’s
Simple DB, Google’s BigTable [9], and Microsoft
SQL Server Data Services.
Architecturally, DryadLINQ is just an application
running on top of Dryad and generating distributed
Dryad jobs. People can envisage making it
interoperate with any of these storage layers.
5. Conclusions
This paper shows the scalability of DryadLINQ by
applying DryadLINQ to a series of scalable
applications with unique requirements. The
applications range from simple map-only operations
such as CAP3 to multiple stages of MapReduce jobs in
CloudBurst. This paper showed that all these
applications can be implemented using the DAG based
programming model of DryadLINQ, and their
performances are comparable to the same applications
developed by using Hadoop.
References
[1] X. Huang and A. Madan, “CAP3: A DNA
Sequence Assembly Program,” Genome Research, vol.
9, no. 9, pp. 868-877, 1999
[2] M. Schatz, “CloudBurst: highly sensitive read
mapping with MapReduce”, Bioinformatics. 2009
June 1; 25(11): 1363-1369.
[3] J. Ekanayake and S. Pallickara, “ MapReduce for
Data Intensive Scientific Analysis,” Fourth IEEE
International Conference on eScience, 2008, pp.277284.
[4] Apache Hadoop, http://hadoop.apache.org/core/
[5] ISARD, M., BUDIU, M., YU, Y., BIRRELL, A.,
AND FET-TERLY, D. Dryad: Distributed dataparallel programs from sequential building blocks. In
Proceedings of European Conference on Computer
Systems (EuroSys), 2007.
[6] CHU, L., TANG, H., YANG, T., AND SHEN, K.
Optimizing data aggregation for clusterbased Internet services. In Symposium on
Principles and practice of parallel
programming (PPoPP), 2003.
[7] DEAN, J., AND GHEMAWAT, S. MapReduce:
Simplified data processing on large clusters.
th
In Proceedings of the 6 Symposium on
Operating Systems Design and Implementation
(OSDI), 2004.
[8] BECK, M., DONGARRA, J., AND PLANK, J. S.
NetSolve/D: A massively parallel grid
execution system for scalable data intensive
collaboration. In International Parallel and
Distributed Processing Symposium (IPDPS),
2005.
[9] CHANG, F., DEAN, J., GHEMAWAT, S., HSIEH,
W. C., WAL- LACH, D. A., BURROWS, M.,
CHANDRA, T., FIKES, A., AND GRUBER, R. E.
BigTable: A distributed storage system for
structured data. In Symposium on Operating
System Design and Implementation (OSDI),
2006.











