ddi12423-sup-0001-AppendixS1-S5
advertisement
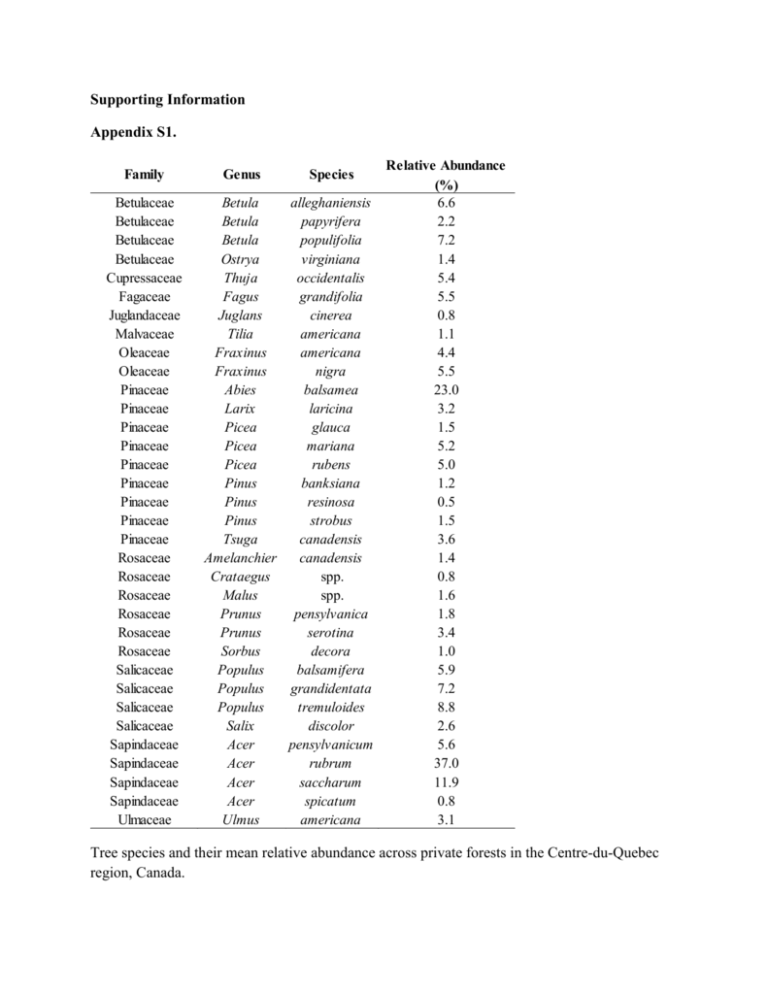
Supporting Information Appendix S1. Family Betulaceae Betulaceae Betulaceae Betulaceae Cupressaceae Fagaceae Juglandaceae Malvaceae Oleaceae Oleaceae Pinaceae Pinaceae Pinaceae Pinaceae Pinaceae Pinaceae Pinaceae Pinaceae Pinaceae Rosaceae Rosaceae Rosaceae Rosaceae Rosaceae Rosaceae Salicaceae Salicaceae Salicaceae Salicaceae Sapindaceae Sapindaceae Sapindaceae Sapindaceae Ulmaceae Relative Abundance (%) Betula alleghaniensis 6.6 Betula papyrifera 2.2 Betula populifolia 7.2 Ostrya virginiana 1.4 Thuja occidentalis 5.4 Fagus grandifolia 5.5 Juglans cinerea 0.8 Tilia americana 1.1 Fraxinus americana 4.4 Fraxinus nigra 5.5 Abies balsamea 23.0 Larix laricina 3.2 Picea glauca 1.5 Picea mariana 5.2 Picea rubens 5.0 Pinus banksiana 1.2 Pinus resinosa 0.5 Pinus strobus 1.5 Tsuga canadensis 3.6 Amelanchier canadensis 1.4 Crataegus spp. 0.8 Malus spp. 1.6 Prunus pensylvanica 1.8 Prunus serotina 3.4 Sorbus decora 1.0 Populus balsamifera 5.9 Populus grandidentata 7.2 Populus tremuloides 8.8 Salix discolor 2.6 Acer pensylvanicum 5.6 Acer rubrum 37.0 Acer saccharum 11.9 Acer spicatum 0.8 Ulmus americana 3.1 Genus Species Tree species and their mean relative abundance across private forests in the Centre-du-Quebec region, Canada. Appendix S2. Responses to Environmental Change Data Type Drought tolerance Climate response index 1-4 Shade tolerance Climate response index 0.98 - 5.01 Water-logging tolerance Climate response index 1 - 3.88 Variable Units Range / Categories Data Source Plant-Environment Traits Niinemets & Valladares 2006 Niinemets & Valladares 2006 Niinemets & Valladares 2006 Life History Traits Maximum height Wood density Response to disturbance Climate response & Response to disturbance numeric cm 500 - 6700 Aubin et al. 2012 numeric g cm -3 0.29 - 0.66 Miles and Smith 2009 Reproduction and Dispersal Traits Mode of Reproduction Response to disturbance nominal Seed mass Response to disturbance numeric Seed dispersal vector Response to disturbance multi-choice nominal Description of functional traits used in the present study. mostly seed, mostly vegetative, only seed g Aubin et al. 2012 Aubin et al. 2012 gravity, endozoochorous, exozoochorous, bird, wind, water, human Aubin et al. 2012 Appendix S3. Sensitivity of relationships between functional response diversity (FD) and community-weighted means (CWMs) of individual traits to trait removal. Relative change in FD was calculated as: 𝐹𝐷−1 −𝐹𝐷 𝐹𝐷 Where FD-1 is FD calculated without the target functional trait and FD is calculating using all traits. Spearman correlations were calculated for CWMs of individual traits and FD-1 Functional Traits T Drought T Shade T Water WD HT SM R Veg R M. Seed R O. Seed D Self D Animal D Exo-animal D Bird D Wind D Water D Human Correlation with FD (all traits) r -0.25 0.10 -0.42 -0.60 -0.27 -0.44 0.26 -0.35 0.21 0.08 0.49 -0.17 0.07 -0.06 -0.52 0.14 P 0.108 0.526 0.005 0.000 0.088 0.003 0.097 0.023 0.191 0.609 0.001 0.272 0.670 0.716 0.000 0.393 Correlation with FD-1 (1 trait removed) r P -0.20 0.207 0.10 0.512 -0.44 0.003 -0.57 0.000 -0.27 0.087 -0.45 0.003 0.05 0.770 -0.11 0.476 0.17 0.271 0.07 0.637 0.40 0.009 -0.17 0.272 0.05 0.773 0.00 0.986 -0.51 0.001 0.14 0.393 Relative Change in FD ( 1 trait removed) 0.01 0.00 -0.03 0.13 0.08 0.07 -0.12 -0.12 -0.12 0.01 0.00 0.00 0.11 0.16 -0.01 0.00 T Drought is drought tolerance, T Shade is shade tolerance, T Water is waterlogging tolerance, WD is wood density, HT is maximum height, SM is seed mass, R Veg. is vegetative reproduction, R M. Seed is reproduction mostly by seed, R O. Seed is reproduction only by seed, D Self is unassisted dispersal, D Animal is endo-zoochorous dispersal by animals, D ExoAnimal is exo-zoochorous dispersal by animals , D Bird is endo-zoochorous dispersal by birds, D Wind is wind dispersal, D Water is water dispersal, and D Human is human-assisted dispersal. Appendix S4. Extended description of estimating functional diversity-area relationships. To generate FD – area curves for both forest types, we created simulated communities by combining species composition data from s randomly selected study sites for all sample sizes (i.e. for s =1, the simulated community is composed of 1 study site, for s = 2, of two study sites, etc.) with replacement 1,000 times (Oksanen, 2007). For each simulated community, FD was calculated and its area was obtained by summing the areas of the selected sites. Simulated data, averaged for each sample size, were then used for model fitting using a normal distribution and a power function. We used a normal distribution for the likelihood function and tested a power function and a Michaelis-Menten model for functional diversity-area relationships: (Eq. 1) 𝑓(𝑥) = 𝑎 ∗ 𝑥 𝑏 (Eq. 2) 𝑓(𝑥) = (𝑎∗𝑥) 𝑎 𝑠 ( ) +𝑥 , where x is the dependent variable and a, b, and s are model parameters. Maximum-likelihood estimates of model parameters were determined using 1 million cycles of a global optimization algorithm (simulated annealing algorithm in the ‘likelihood’ package). Two different model types, Michaelis-Menten and power functions, were fitted initially and the most parsimonious was selected using AICc. For deciduous forests, a power function was applied to patches with areas between 8.83 and 194 ha and for mixed forests, to patches with areas between 8.57 and 171 ha. Patches with areas outside of these ranges were assigned maximum and minimum predicted values. Appendix S4 (continued). Model selection for functional diversity – area relationships. Variables Dependent Area (ha) Area (ha) Independent FD FD Forest Type Model Type Model Fit 2 Deciduous Mixed power power Model Parameters ΔAICc R RMSE a b 50.86 97.4% 0.004 0.31 0.07 54.45 98.2% 0.002 0.32 0.04 s SD 0.004 0.002 ΔAICc is the difference between the best fit and the competing model and RMSE is root mean squared error. A,b, and s are model parameters and SD is standard deviation. Functional diversity accumulation curves and model fit information for mixed (dotted line) and deciduous (solid line) private forests in the Centre-du-Quebec region, Canada Appendix S5. Mathematical expressions for dPCconnectork , dPCintrak and dPCfluxk In all expressions n denotes the total number of patches in the landscape and fi is the FD of patch i. 𝑑𝑃𝐶𝑐𝑜𝑛𝑛𝑒𝑐𝑡𝑜𝑟𝑘 = 100 𝑃𝐶 ∗𝑘 ∗ ∑𝑛𝑖=1 ∑𝑛𝑗=1 𝑓𝑖 𝑓𝑗 (𝑝𝑖𝑗 − 𝑝𝑖𝑗_𝑟𝑒𝑚𝑜𝑣𝑒,𝑘, ) 𝑖, 𝑗 ≠ 𝑘 and 𝑖 ≠ 𝑗 (1) ∗𝑘 Where 𝑝𝑖𝑗 is the maximum dispersal probability between patches i and j in the intact network. The ∗ most probable path between i and j may include patch k. On the other hand, 𝑝𝑖𝑗_𝑟𝑒𝑚𝑜𝑣𝑒,𝑘, is the maximum dispersal probability between patches i and j in the landscape-network where patch k has been removed. Note that dPCconnectork is independent of fk. 𝑑𝑃𝐶𝑖𝑛𝑡𝑟𝑎𝑘 = 𝑑𝑃𝐶𝑓𝑙𝑢𝑥𝑘 = 100 𝑃𝐶 100 𝑃𝐶 𝑓𝑘2 ∗ ∗ 2 ∑𝑛𝑖=1 𝑓𝑖 𝑓𝑘 𝑝𝑖𝑘 (2) 𝑖≠𝑘 (3) ∗ Where 𝑝𝑖𝑘 is the maximum probability of dispersal between i and k in the intact network (before the removal of k).