Extended Prediction, Biodiversity Gradients
advertisement

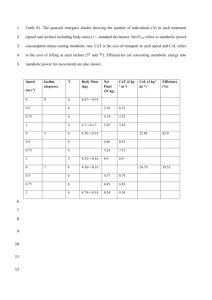
Supplemental Information: Testing the metabolic theory of ecology Level 4 - Evaluating MTE’s Extended predictions Here we consider three areas that have built on the basic MTE foundation to yield additional predictions. The three areas are ontogenetic growth, size-frequency distributions and biodiversity gradients. Extended Prediction, Ontogenetic Growth: One of the earliest extensions of Model A was the development of a model of ontogenetic growth (West et al. 2001). As in the case of the scaling of metabolic rate, WBE were building on a field with a rich modelling history (von Bertalanffy 1957; Ricklefs 1969; Reiss 1989; Kooijman 1993). WBE assume that the total metabolic rate of an organism (B) is the sum of the metabolic expenditure devoted to growth (i.e., the synthesis of new biomass) and the metabolic expenditure devoted to maintaining existing biomass, B Em dm Bm m dt (1) where Em is the energy required to synthesize a unit of biomass (e.g., in J g-1), and Bm is the power required to maintain a unit of biomass (e.g., in W g-1) , and m is the total mass of the organism at time t. Further, they assumed the ¾-power allometry for total metabolic rate, i.e., B = B0 m3/4 where B0 is a coefficient that depends on taxa, temperature, and metabolic state. Solving for the rate of growth in mass yields the relationship 3 dm am 4 bm dt , (2) B where m is the mass of the growing animal, a B0 and b m . Thus, a is the ratio of the Em Em metabolic scaling coefficient (B0) to the energetic cost of biomass synthesis, while b is the energy to create it. ratio of the power to maintain tissue relative to the Interestingly, and perhaps not surprisingly, this model produces a similar-shaped prediction as earlier efforts in modelling the generally sigmoid pattern of ontogenetic growth trajectories (von Bertalanffy 1957; Ricklefs 1969; Reiss 1989), but differs from its predecessors in its underlying assumptions and in explicitly linking the parameters of the equation to basic cellular and tissue properties. As a result, several authors noted that while the fits of the model to data from a wide variety of animal species were quite impressive, support for the details of the model, and especially for the universality of the ¾ metabolic exponent, could not be concluded from statistical fits of growth trajectories alone (Banavar et al. 2002; Ricklefs 2003). In the case of the original growth model, the authors rely on the assumption that the ¾-power scaling across species applies equally well to developmental size changes. Glazier questioned this assumption (Glazier 2005), and while the overall average of the scaling exponent is around ¾, especially for organisms growing over a substantial size range (Moses et al. 2008), there is considerable variation present across intraspecific metabolic scaling relationships (von Bertalanffy 1957; Glazier 2005). Later versions of the model have been generalized to incorporate both variation in the metabolic scaling exponent (Moses et al. 2008) and a more thorough partitioning of energy assimilation and allocation (Hou et al. 2008) that together may provide more flexible and robust predictions. Summary: While the simplicity of the original WBE growth model combines MTE with a theory of ontogenetic growth, it has limitations that are being addressed with additional research. The considerable phylogenetic, ecological, and ontogenetic variation present across intraspecific metabolic scaling relationships suggests that applying the MTE to patterns of ontogenetic growth will require a flexible theory that relaxes many of the original assumptions. Further ontogenetic studies of growth and metabolism in model organisms (Yagi et al. 2010; Sears et al. 2012) will help to provide stronger tests of the underlying model assumptions and to adapt the generalized theory to more specific biological contexts. Extended prediction, size-frequency distributions: Enquist et al. (1998) applied MTE to the size-abundance relationship within plant communities. By assuming resource limited growth, a steady state in resource flux, and scaling of metabolic rate with body mass to the ¾ power, Enquist et al.(1998) predicted that maximum abundance should scale with mean body mass to the -3/4 power. These authors found good empirical support for this prediction and indirectly tested their assumed size scaling of metabolism. MTE has also been applied to size-abundance relationships within mixed-age, mixed-species forest stands (Enquist & Niklas 2001) by estimating the scaling exponent for a large number of within-stand sizeabundance relationships. Later models derived that the key prediction is that abundance declines with stem diameter to the -2 power. This prediction actually corresponds to a decline of abundance with mean body mass to the -7/8 power, and reasonable support was found for this prediction (see also, Stegen & White 2008; Enquist et al. 2009; West et al. 2009). Summary: Reanalysis of the global data from Enquist and Niklas (2001) and several tropical sites (Muller-Landau et al. 2006) has revealed that size-frequency distributions are often not straight lines on log-log axes (Coomes et al. 2003) and show extensive covariation in both the scaling exponent and allometric constant of forest self-thinning (Niklas et al. 2003). Furthermore, size-distributions within populations undergoing stand development are better described by Weibull distributions than power functions (Mohler et al. 1978; Muller-Landau et al. 2006; Coomes & Allen 2007). Hence, the use of mechanistic models that incorporate species level information on allometric dependencies of growth and mortality may provide a potential route to develop simplified models that can explain observed forest data (e.g., see the work of Purves et al. (2008)). Extended Prediction, Biodiversity Gradients: Metabolic influence over species packing. The first formal link between MTE and diversity gradients was developed in Allen et al. (2002). These authors started with the (Boltzmann) assumption for metabolic rate dependence on temperature and predicted that the number of individuals of a given species within a given area will decrease exponentially with temperature. Three further assumptions underlie this approach: namely that the limiting resource supply, population-level energy use per area, and the number of individuals per area, all do not vary with temperature. The model then predicts that at higher temperatures each species is represented by fewer individuals, but the whole community contains the same number of individuals, so that the number of species must increase with temperature. Allen et al. (2002) assume that limiting resource supply does not vary with temperature so that energy use across all individuals at the population- or community-level should not vary with temperature. However, the model also assumes that total community abundance and average body size do not vary with temperature. In this case community-level energy use must increase with temperature. As such, the model does not appear to be internally consistent (Storch 2003; Stegen et al. 2009). However as Allen et al. (2002) point out, the model may be robust to violations of some assumptions, an issue that warrants further investigation. Metabolic influence over mutation and speciation rates. A separate line of enquiry explores the influence of metabolic rates on rates of genetic mutation (Gillooly et al. 2005; Gillooly et al. 2007) and speciation (Allen et al. 2006). Allen et al. (2006) assume the following: that both the number of mutations, and the number of generations per time increase with metabolic rate, and thus both increase exponentially with temperature; that the number of genetic changes required for speciation, effective population size, and selection strength are all independent of temperature; that speciation is proportional to overall mutation rate, avoiding the need for a causal link with species ecology. Combining these assumptions leads to the prediction that speciation rate will increase exponentially with temperature. Correlations of speciation rate with temperature can thus be used to support the models predictions, but do not directly validate the models’ underlying assumptions or mechanism (Davies et al. 2004; Stegen et al. 2009). Some assumptions have been tested (e.g. the temperature dependence of mutation rate, Gillooly et al. 2005), while others are yet to be directly evaluated (e.g. the temperature independence of effective population size, Stegen et al. 2009). Summary: Meta-analyses of global datasets for plant, invertebrate and ectothermic vertebrate groups uncovered a broad range of relationships between species richness and environmental temperature (Hawkins et al. 2007). The majority of these patterns depart significantly from the pattern predicted by Allen et al. (2002), suggesting that there is little support for current extensions of MTE as a predictor of global diversity gradients and a need for further modelling (e.g. Stegen et al. 2012). Literature Cited Allen A.P., Brown J.H. & Gillooly J.F. (2002). Global biodiversity, biochemical kinetics, and the energetic equivalence rule. Science, 297, 1545-1548. Allen A.P., Gillooly J.F., Savage V.M. & Brown J.H. (2006). Kinetic effects of temperature on rates of genetic divergence and speciation. Proceedings of the National Academy of Science of the United States of America, 103, 9130-9135. Banavar J.R., Damuth J., Maritan A. & Rinaldo A. (2002). Supply-demand balance and metabolic scaling. Proceedings of the National Academy of Science of the United States of America, 99, 10506-10509. Coomes D.A. & Allen R.B. (2007). Mortality and tree-size distributions in natural mixed-age forests. J. Ecol., 95, 27-40. Coomes D.A., Duncan R.P., Allen R.B. & Truscott J. (2003). Disturbances prevent stem size-density distributions in natural forests from following scaling relationships. Ecol. Lett., 6, 980-989. Davies T.J., Savolainen V., Chase M.W., Moat J. & Barraclough T.G. (2004). Environmental energy and evolutionary rates in flowering plants. Proc. R. Soc. Lond. Ser. B-Biol. Sci., 271, 2195-2200. Enquist B.J., Brown J.H. & West G.B. (1998). Allometric scaling of plant energetics and population density. Nature, 395(6698), 163-165. Enquist B.J. & Niklas K.J. (2001). Invariant scaling relations across tree-dominated communities. Nature, 410, 655-660. Enquist B.J., West G.B. & Brown J.H. (2009). Extensions and evaluations of a general quantitative theory of forest structure and dynamics. Proceedings of the National Academy of Sciences, 106, 7046-7051. Gillooly J.F., Allen A.P., West G.B. & Brown J.H. (2005). The rate of DNA evolution: Effects of body size and temperature on the molecular clock. Proc. Natl. Acad. Sci. U. S. A., 102, 140-145. Gillooly J.F., McCoy M.W. & Allen A.P. (2007). Effects of metabolic rate on protein evolution. Biology Letters, 3, 655-659. Glazier D.S. (2005). Beyond the '3/4-power law': variation in the intra- and interspecific scaling of metabolic rate in animals. Biol. Rev., 80, 611-662. Hou C., Zou W., Moses M.E., Woodruff W.H., Brown J.H. & West G.B. (2008). Energy Uptake and Allocation During Ontogeny. Science, 322, 736-739. Kooijman S.A.L.M. (1993). Dynamic Energy Budgets in Biological Systems. Cambridge University Press, Cambridge. Mohler C.L., Marks P.L. & Sprugel D.G. (1978). Stand structure and allometry of trees during selfthinning of pure stands. J. Ecol., 66, 599. Moses M.E., Hou C., Woodruff W.H., West G.B., Nekola J.C., Zuo W. & Brown J.H. (2008). Revisiting a model of ontogenetic growth: Estimating model parameters from theory and data. Am. Nat., 171, 632-645. Muller-Landau H.C., Condit R.S., Harms K.E., Marks C.O., Thomas S.C., Bunyavejchewin S., Chuyong G., Co L., Davies S., Foster R., Gunatilleke S., Gunatilleke N., Hart T., Hubbell S.P., Itoh A., Kassim A.R., Kenfack D., LaFrankie J.V., Lagunzad D., Lee H.S., Losos E., Makana J.R., Ohkubo T., Samper C., Sukumar R., Sun I.F., Supardi N.M.N., Tan S., Thomas D., Thompson J., Valencia R., Vallejo M.I., Munoz G.V., Yamakura T., Zimmerman J.K., Dattaraja H.S., Esufali S., Hall P., He F.L., Hernandez C., Kiratiprayoon S., Suresh H.S., Wills C. & Ashton P. (2006). Comparing tropical forest tree size distributions with the predictions of metabolic ecology and equilibrium models. Ecol. Lett., 9, 589-602. Niklas K.J., Midgley J.J. & Enquist B.J. (2003). A general model for mass-growth-density relations across tree- dominated communities. Evolutionary Ecology Research, 5, 459-468. Reiss M. (1989). Plant resource allocation. TREE, 4, 379-380. Ricklefs R. (2003). Is rate of ontogenetic growth constrained by resource supply or tissue growth potential? A reply to West et al.'s model. Funct. Ecol., 17, 384-393. Ricklefs R.E. (1969). Preliminary models for growth rates in altricial birds. Ecology, 50, 1031-1039. Sears K.E., Kerkhoff A.J., Messerman A. & Itagaki H. (2012). Ontogenetic Scaling of Metabolism, Growth, and Assimilation: Testing Metabolic Scaling Theory with Manduca sexta Larvae. Physiol. Biochem. Zool., 85, 159-173. Stegen J.C., Enquist B.J. & Ferriere R. (2009). Advancing the metabolic theory of biodiversity. Ecol. Lett., 12, 1001-1015. Stegen J.C., Ferriere R. & Enquist B.J. (2012). Evolving ecological networks and the emergence of biodiversity patterns across temperature gradients. Proc R Soc Lond Biol, 279, 1051-1060. Stegen J.C. & White E.P. (2008). On the relationship between mass and diameter distributions in tree communities. Ecol. Lett., 11, 1287-1293. Storch D. (2003). Comment on "Global biodiversity, biochemical kinetics, and the energeticequivalence rule". Science, 299, 346b. von Bertalanffy L. (1957). Quantitative laws in metabolism and growth. Quarterly Review of Biology, 32, 217-231. West G.B., Brown J.H. & Enquist B.J. (2001). A general model for ontogenetic growth. Nature, 413, 628-631. West G.B., Enquist B.J. & Brown J.H. (2009). A general quantitative theory of forest structure and dynamics. Proceedings of the National Academy of Sciences, 106, 7040-7045. Yagi M., Kanda T., Takeda T., Ishimatsu A. & Oikawa S. (2010). Ontogenetic phase shifts in metabolism: links to development and anti-predator adaptation. Proceedings Of The Royal Society B-Biological Sciences, 277, 2793-2801.