tpj13042-sup-0012
advertisement

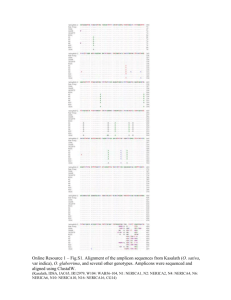
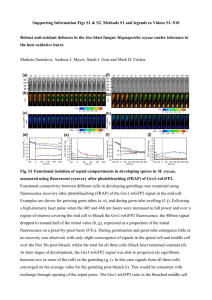
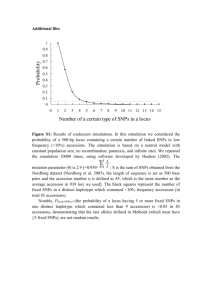
Supporting Information Legends Supplementary Figures Figure S1. Sequence alignment of pattern 1, 2 and 3 OsSWEET14 promoters regions. The 334-bp OsSWEET14 promoter sequences obtained from the O. sativa ssp. japonica cultivar Nipponbare (pattern 1), O. sativa spp. indica cultivar IR8 (336) (pattern 1), O. longistaminata accession LL109 (pattern 1), O. barthii accession 554W1 (pattern 1), O. barthii accessions 530A1, 534A1 and 536A1 (pattern 2), O. glaberrima accession CG14 (pattern 3) and O. barthii accession 541W1 (pattern 3) were aligned using ClustalW. For pattern-3 accessions, PCR amplicons were subcloned into the pGEM-T plasmid and sequences corresponding to each of the two promoter variants are presented (#1, with deletion, #2, without deletion). Blue, red and black fonts indicate pattern 1, 2 and 3 accessions, respectively. The TalC (1), ArtTAL14-2 (2), AvrXa7 (3) and Tal5 (2) EBEs are respectively depicted in bold, framed in black, underlined and highlighted in grey, respectively. The 5’ terminal nucleotide of each target box is indicated in white font. Figure S2. xa41(t) defeats X. oryzae pv. oryzae strains of diverse genetic and geographic origins. Minimum spanning tree (MST) of a subset of X. oryzae pv. oryzae strains available at the IRD collection using Bionumerics 7.1 (4). Colored circles indicate strains that were used to infiltrate leaves of two week-old O. barthii xa41(t) 536A1 and Xa41(t) 554W1 accessions. Color is attributed according to the country of origin of the strain as indicated on the right. A grey halo depicts strains that are defeated by xa41(t). Symptoms were analyzed 5 days after inoculation. This experiment was reproduced three times with similar results. Figure S3. xa41(t)-mediated resistance is associated with a loss of OsSWEET14 induction. Leaves of O. barthii accessions 536A1 and 554W1 respectively carrying xa41(t) and Xa41(t) were infiltrated with X. oryzae pv. oryzae strains BAI3, BAI4, PXO86 and NXO37. OsSWEET14 transcript abundances were determined 2 days after inoculation by quantitative reverse transcription polymerase chain reaction (qRT-PCR). The bars represent three biological replicates. The fold change was calculated in comparison to leaves treated with water. Actin was used as a reference gene to normalize cDNA amounts. Error bars represent SD (n=3). An asterisk indicates a significant difference in ANOVA analysis at P < 0.05. Indications below refer to the ability of strains to cause BLB symptoms (+) or not (-) in rice accessions carrying xa41(t) (Table 1). Figure S4. Segregation analysis of xa41(t) in pattern 3 accessions using a restriction fragment length polymorphism/polymerase chain reaction approach. (A) Schematic of the OsSWEET14 promoter region depicted with BstNI restriction recognition sites, the size of BstNI RFLP/PCR products, and the EBEs of TalC, AvrXa7 and Tal5. The 18-bp sequence deleted in xa41(t) accessions is indicated as a dashed box (). Since it bears a BstNI recognition site, it is possible to discriminate between OsSWEET14 promoters carrying or not the 18-bp deletion according to the sizes of the restriction fragments. As indicated, four RFLP/PCR products with a size of 120bp, 98bp, 268bp and 21bp are expected in absence of the 18-bp deletion, while products of 120bp, 348bp and 21bp are estimated for promoters with the deletion. (B) A 507-bp fragment of the OsSWEET14 promoter was amplified using primers pSWEET14_Fw and pSWEET14_Rev and genomic DNA of O. sativa cv. Nipponbare (1), O. barthii accessions 554W1 (2), 527W1 (3), 536A1 (4), 534A1 (5) and 530W1 (6), O. glaberrima cv. CG14 (7), O. glaberrima cv. 5681 (8) and O. barthii acc. 541W1 (9). Upon digestion with BstNI, RFLP/PCR products were analyzed by gel electrophoresis. As expected pattern 1 (lanes 1-3), pattern 2 (lanes 4-6) and pattern 3 (lanes 7-9) accessions are respectively characterized by restriction fragments of 268bp, 348bp and 268/348bp. (C) Segregation analysis of xa41(t) in pattern 3 accessions. Twenty-eight progeny individuals of O. glaberrima cultivars TOG5681, TOG5418 and TOG5984, and O. barthii accessions 541W1, 535A1 and 535W1 were genotyped by RFLP/PCR. For each progeny, results were identical among the 28 tested individuals, six of which are presented here for O. glaberrima cv. CG14, (lanes 13-18). As control, 6 progeny individuals of O. barthii 554W1 (pattern 1, lanes 1-6) and O. barthii 536A1 (pattern 2, lanes 7-12) were genotyped as well.. Lanes 1 to 12 were run on the same gel, but some lanes were removed for presentational purposes, as indicated by the white lines. Figure S5. xa41(t) is functional in O. glaberrima. Leaves of two week-old O. barthii 554W1 (pattern 1) and 536A1 (pattern 2), O. glaberrima cultivars CG14, TOG5681 and 106, and O. barthii accession 541W1 (pattern 3), were infiltrated with water, X. oryzae pv. oryzae strain BAI3, and X. oryzae pv. oryzae BAI3ΔtalC derivatives carrying an empty vector (ev) or plasmids containing the TAL effector genes tal5, avrXa7, talC or artTAL14-2. Symptoms were photographed 5 days after inoculation. This experiment was reproduced three times with similar results. Supplementary Tables Table S1. Oryza spp. accessions analyzed for polymorphism of the OsSWEET14 promoter. Table S2. The pattern-2 accessions 536A1, 534A1 and 530A1 are resistant to Xoo strains relying on tal5 and avrXa7 for virulence. Table S3. talC and artTAL14-2 confer a gain of virulence to PXO86 on xa41(t) rice. Table S4. Xanthomonas oryzae pv. oryzae strains used to evaluate xa41(t) resistance spectrum. Table S5. Primers used in this study. Supplementary Text Text S1. OsSWEET14 and OsSWEET14_trunc consensus sequences obtained upon PacBio sequencing of Oryza glaberrima cultivars CG14 and TOG5681. The consensus sequences are depicted with the promoter region highlighted in grey, the 18-bp deletion in pink, the predicted exons and introns in blue and yellow respectively (6), while the predicted start codon of OsSWEET14 is indicated in bold italic font. The duplicated sequence located upstream of the promoter is highlighted in green.