Table S1 Specimen numbers, localities and GenBank
advertisement
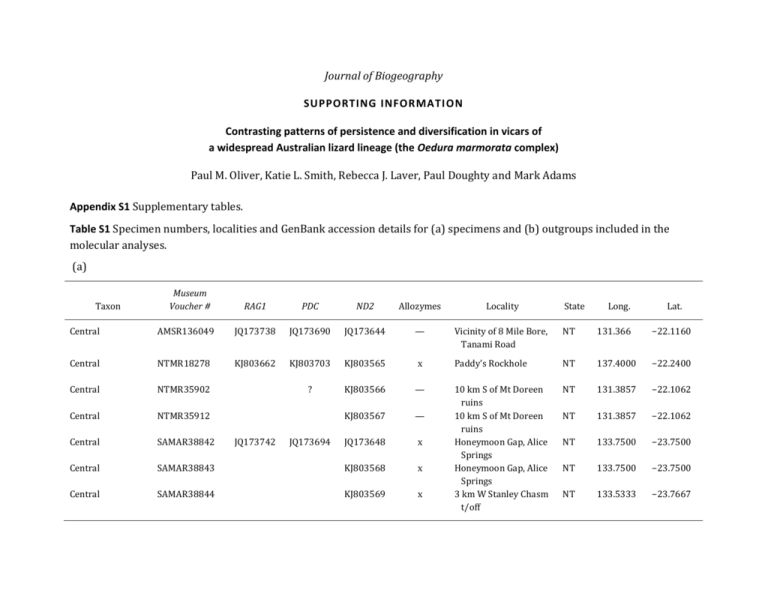
Journal of Biogeography SUPPORTING INFORMATI ON Contrasting patterns of persistence and diversification in vicars of a widespread Australian lizard lineage (the Oedura marmorata complex) Paul M. Oliver, Katie L. Smith, Rebecca J. Laver, Paul Doughty and Mark Adams Appendix S1 Supplementary tables. Table S1 Specimen numbers, localities and GenBank accession details for (a) specimens and (b) outgroups included in the molecular analyses. (a) Taxon Museum Voucher # RAG1 PDC ND2 Allozymes Locality State Long. Lat. Central AMSR136049 JQ173738 JQ173690 JQ173644 — Vicinity of 8 Mile Bore, Tanami Road NT 131.366 −22.1160 Central NTMR18278 KJ803662 KJ803703 KJ803565 x Paddy’s Rockhole NT 137.4000 −22.2400 Central NTMR35902 ? KJ803566 — NT 131.3857 −22.1062 Central NTMR35912 KJ803567 — NT 131.3857 −22.1062 Central SAMAR38842 JQ173648 x NT 133.7500 −23.7500 Central SAMAR38843 KJ803568 x NT 133.7500 −23.7500 Central SAMAR38844 KJ803569 x 10 km S of Mt Doreen ruins 10 km S of Mt Doreen ruins Honeymoon Gap, Alice Springs Honeymoon Gap, Alice Springs 3 km W Stanley Chasm t/off NT 133.5333 −23.7667 JQ173742 JQ173694 Taxon Museum Voucher # RAG1 PDC ND2 Allozymes Locality 3 km W Stanley Chasm t/off Emily Gap Bourke, west of Warego River Bridge 0.5 km S Angepena HS Oakbank Station 30 km Eof Noonbah Station 85 km W Windorah 2.7 km W Lance Bore Narrina Stn 6 km ESE Blackwater Springs,Oratunga W of Bellbird Campsite, Gluepot Reserve 9 km N NSW/QLD border on Mitchell Highway 0.5 km W Ballards tank, Plevna Downs Musselbrook Reservoir Lawn Hill NP Mount Isa McArthur R. Stn. Urapunga Station Victoria R. Gregory NP. 3 km S Katherine Marchinbar Is, (North) Central SAMAR38845 KJ803570 x Central Eastern SAMAR65905 AMSR138446 JQ173691 KJ803571 JQ173645 x — Eastern Eastern Eastern SAMAR38901 SAMAR41324 SAMAR42883 KJ803663 KJ803664 KJ803704 KJ803705 — KJ803573 KJ803574 x x x Eastern Eastern SAMAR42913 SAMAR52203 JQ173743 JQ173695 KJ803575 JQ173649 x — Eastern SAMAR52333 KJ803576 x Eastern SAMAR54300 KJ803665 KJ803706 KJ803577 x Eastern SAMAR55902 KJ803666 KJ803707 KJ803578 x Eastern ABTC113886 KJ803572 x Gulf1 Gulf1 Gulf1 Gulf2 North1 North1 North1 North1 NTMR21288 SAMAR34209 SAMAR35425 SAMAR34188 NTMR13222 NTMR13295 NTMR13619 NTMR19029 KJ803591 KJ803593 KJ803594 KJ803592 KJ803596 KJ803597 JQ173647 KJ803598 x x x x x x — x JQ173739 KJ803670 KJ803711 KJ803672 KJ803671 KJ803673 KJ803674 JQ173741 KJ803675 KJ803713 KJ803712 KJ803714 KJ803715 JQ173693 KJ803716 State Long. Lat. NT 133.5333 −23.7667 NT NSW 133.9506 145.3500 −23.7397 −30.0000 SA SA QLD 138.8500 140.6444 143.4167 −30.5750 −33.1278 −24.1167 QLD SA 141.8333 138.7730 −25.3500 −30.9567 SA 138.8836 −31.0972 SA 140.1631 −33.7028 QLD 145.7278 −28.9603 Qld 142.5628 −26.7394 QLD Qld QLD NT NT NT NT NT 138.3133 138.5000 139.4833 135.8500 134.5700 130.5006 132.27 136.6986 −18.4000 −18.5830 −20.7333 −16.6667 −14.5100 −15.9433 −14.47 −11.1978 Taxon Museum Voucher # RAG1 PDC ND2 Allozymes Locality State Long. Lat. North1 NTMR19030 KJ803676 KJ803717 KJ803599 x Marchinbar Is, (North) NT 136.6986 −11.1978 North1 NTMR22774 KJ803677 KJ803718 KJ803600 x NT 136.4222 −11.8797 North1 North1 North1 North1 North2 North2 North3 North3 North4 Oedura filicipoda Oedura filicipoda NTMR36709 NTMR36717 NTMR36731 NTMR36746 SAMAR34156 SAMAR34158 NTMR22104 NTMR22183 NTMR22444 AMSR126183 WAMR138874 KJ803595 KJ803601 KJ803602 KJ803603 KJ803604 KJ803605 KJ803606 KJ803607 KJ803608 JQ173636 KJ803581 — — — — x x x x x — — NT NT NT NT NT NT NT NT NT WA WA 130.7866 134.3640 134.1611 134.3430 132.2583 132.2583 130.8167 130.8967 135.3311 125.7160 125.5069 -14.0739 −14.1780 −14.1529 −14.1328 −14.4583 −14.4583 −12.5167 −13.4086 −15.7806 −14.8160 −14.9875 Oedura filicipoda Oedura filicipoda WAMR167805 WAMR171552 KJ803579 KJ803580 — — WA WA 125.7322 125.2575 −14.6733 −15.7613 Oedura gemmata NMVD72584 KJ803582 — NT 133.8125 −12.1883 Oedura gemmata NMVD72598 KJ803583 — NT 133.8125 −12.1883 Oedura gemmata NMVD72599 KJ803584 — NT 133.8125 −12.1883 Oedura gemmata NMVD72600 KJ803585 — NT 133.8125 −12.1883 Oedura gemmata NMVD72621 KJ803586 — English Company Isles, Astell Is Fish River Wongalarra Wongalarra Wongalarra 1 km N Katherine 1 km N Katherine Cox Peninsula Rd Litchfield NP Limmen Gate NP Little Mertens Falls Donkin’s Hill, Mitchell Plateau Mitchell Plateau Prince Regent Nature Reserve Near Kikiyown (also called ‘Kikikyon’) Near Kikiyown (also called ‘Kikikyon’) Near Kikiyown (also called ‘Kikikyon’) Near Kikiyown (also called ‘Kikikyon’) Near Kikiyown (also called ‘Kikikyon’) NT 133.8125 −12.1883 KJ803678 KJ803679 KJ803680 KJ803681 KJ803719 KJ803720 KJ803721 KJ803722 KJ803682 JQ173730 KJ803729 KJ803723 JQ173682 KJ803729 Taxon Museum Voucher # RAG1 PDC ND2 Allozymes Locality NT 133.1772 −13.9805 NT 133.1772 −13.9805 NT NT NT WA 132.9788 133.0722 132.4167 125.1830 −13.8617 −12.3762 −13.4333 −16.9830 WA 125.9269 −16.6586 NT WA 129.7500 125.5069 −15.4667 −14.9875 WA WA 128.2867 125.9500 −16.7508 −14.3500 WA WA WA WA WA WA WA WA 128.4117 125.1689 125.5208 125.2000 124.5333 124.5611 123.4714 126.5000 −17.4002 −17.6377 −14.4500 −15.1000 −15.3500 −15.9500 −16.6236 14.8000 WA WA WA WA 127.0539 120.4067 120.4067 120.4067 −15.9839 −24.0536 −24.0536 −24.0536 Oedura gemmata NTMR34986 KJ803708 KJ803708 KJ803587 — Oedura gemmata NTMR34987 KJ803709 KJ803709 KJ803588 — Oedura gemmata Oedura gemmata Oedura gemmata Oedura gracilis NTMR35680 NTMR35753 SAMAR34170 AMSR136067 KJ803710 KJ803710 JQ173731 JQ173732 JQ173683 JQ173684 KJ803589 KJ803590 JQ173637 JQ173638 — — x — Oedura gracilis AMSR140309 JQ173733 JQ173685 JQ173639 — Oedura gracilis Oedura gracilis NTMR13438 WAMR138891 KJ803741 KJ803738 KJ803741 KJ803738 KJ803620 KJ803617 — — Oedura gracilis Oedura gracilis WAMR151005 WAMR151963 KJ803740 KJ803742 KJ803740 KJ803742 KJ803619 KJ803621 — — Oedura gracilis Oedura gracilis Oedura gracilis Oedura gracilis Oedura gracilis Oedura gracilis Oedura gracilis Oedura gracilis WAMR156724 WAMR156728 WAMR164910 WAMR168564 WAMR168565 WAMR171668 WAMR171670 WAMR172341 KJ803730 KJ803731 KJ803732 KJ803733 KJ803734 KJ803735 KJ803736 KJ803737 KJ803730 KJ803731 KJ803732 KJ803733 KJ803734 KJ803735 KJ803736 KJ803737 KJ803609 KJ803610 KJ803611 KJ803612 KJ803613 KJ803614 KJ803615 KJ803616 — — — — — — — — Mountain Valley Station Mountain Valley Station Kakadu National Park Oenpelli, Reservoir UDP Falls Bells Gorge, Bells Crk, Isdell River Manning Gorge, Mt. Barnett Station Bulloo River Station Donkin's Hill, Mitchell Plateau Warmun South West Osborn Island Piccaninny Massif Tunnel Creek Katers Island Boongaree Island Augustus Island Storr Island Lachlan Island Theda Station Oedura gracilis Western Western Western WAMR172865 WAMR102618 WAMR102619 WAMR102622 KJ803739 KJ803739 KJ803618 KJ803622 KJ803623 KJ803624 — x x x Ellenbrae Little Sandy Desert Little Sandy Desert Little Sandy Desert State Long. Lat. Taxon Museum Voucher # RAG1 PDC ND2 Allozymes Locality JQ173744 JQ173696 JQ173650 KJ803625 KJ803626 KJ803627 KJ803628 x x x x x 7 km N Mt Magnet WA 7 km N Mount Magnet 4 km SW Hugh Bluff Balfour Downs NW end of Mount Fraser Virgin Springs, Carnarvon Range Yandicoogina WA Hope Downs Mount Brockman Hope Downs Yandicoogina unknown Mount Brockman, Namuldin 120 km NW Newman 120 km NW Newman 120 km NW Newman Ulongunna Rock Burrup Peninsula Burrup Peninsula Mount Brockman Mount Brockman Mount Brockman Mount Brockman Mount Brockman 40 km SE Pouyouwuncubban Western Western Western Western Western WAMR105965 WAMR106289 WAMR110173 WAMR111891 WAMR114252 Western WAMR119086 KJ803629 x Western Western Western Western Western Western Western WAMR119837 WAMR119991 WAMR119992 WAMR119993 WAMR125106 WAMR126074 WAMR127833 KJ803630 KJ803631 KJ803632 KJ803633 KJ803634 KJ803635 KJ803636 x x x x x x x Western Western Western Western Western Western Western Western Western Western Western Western WAMR129595 WAMR129622 WAMR129635 WAMR132296 WAMR132626 WAMR132633 WAMR135369 WAMR135445 WAMR135447 WAMR135448 WAMR135449 WAMR146593 KJ803637 KJ803638 KJ803639 KJ803640 KJ803641 KJ803642 KJ803643 KJ803644 KJ803645 KJ803646 KJ803647 KJ803648 x x x x x x x x x x x x KJ803724 KJ803724 State Long. Lat. WA WA WA WA WA 117.8494 117.8500 117.6930 120.1000 118.3667 −28.0097 −28.0000 −22.0592 −23.5000 −25.5833 WA 120.7167 −25.1000 WA WA WA WA WA WA WA 120.1800 119.1028 117.3000 119.1167 119.0167 ? 117.3219 −21.3997 −23.0083 −22.4666 −23.0000 −22.7166 ? −22.3105 WA WA WA WA WA WA WA WA WA WA WA WA 118.8833 118.8833 119.0167 117.2331 116.8108 116.8108 117.3219 117.3219 117.3219 117.3219 117.3219 119.0186 −22.9166 −22.9166 −22.9166 −27.1194 −20.5963 −20.5963 −22.3105 −22.3105 −22.3105 −22.3105 −22.3105 −22.1494 Taxon Museum Voucher # Western WAMR146594 Western WAMR154783 Western Western Western Western Western Western Western WAMR154785 WAMR154796 WAMR154797 WAMR157504 WAMR157516 WAMR157595 WAMR160066 Western WAMR160074 Western WAMR165150 Western Western WAMR165241 WAMR165242 RAG1 KJ803725 PDC KJ803725 KJ803726 KJ803726 KJ803727 KJ803727 KJ803728 KJ803728 ND2 Allozymes Locality State Long. Lat. KJ803649 x WA 119.0186 −22.1494 KJ803650 x 40 km SE Pouyouwuncubban Brockman Ridge WA 119.8861 −23.3091 KJ803651 KJ803652 KJ803653 KJ803654 KJ803655 KJ803656 KJ803657 x x x x x x x WA WA WA WA WA WA WA 119.9169 117.4708 117.4708 118.8900 118.8900 118.8606 121.0020 −23.3108 −27.3986 −27.3986 −22.9169 −22.9169 −23.1869 −21.3219 KJ803658 x WA 120.7520 −21.3337 KJ803659 x WA 117.2310 −21.3210 KJ803660 KJ803661 — x Brockman Ridge Walga Rock Walga Rock Packsaddle Range Packsaddle Range West Angelas 58 km ESE Meentheena Outcamp 32.5 km ESE Meentheena Outcamp 1.5 km NNW Python Pool Hanson Cove Burrup Peninsula, Hearson Cove WA WA 116.7989 116.7990 −20.6352 −20.6350 (b) Locality Outgroup species Museum number Crenadactylus ocellatus Diplodactylus conspicillatus Diplodactylus granariensis Diplodactylus ornatus Diplodactylus tessellatus Lucasium maini Lucasium stenodactylum Mokopirirakau granulatus Oedodera marmorata Oedura castelnaui Oedura castelnaui Oedura coggeri SAMA R22245 AMS R158426 AMS R150637 AMS R140546 AMS R143855 AMS150647 AMS R139897 RAH363 AMS R161254 AMS R143917 SAMA R55715 AMS R143918 Amalosia lesueurii Amalosia lesueurii Amalosia lesueurii Oedura monilis Oedura monilis Oedura monilis Amalosia obscura Amalosia obscura Hesperoedura reticulate Amalosia rhombifer Amalosia rhombifer Amalosia rhombifer AMS R152230 AMS R159546 AMS R161008 SAMA R54507 SAMA R54560 AMS R152056 AMS R136124 AMS R136077 SAMA R23035 SAMA R55604 NTMR 22222 SAMA R34513 NT: 10 km S Barrow Ck NSW: Sturt National Park WA: Dedari WA: Denham QLD: 7.9 km SW of Landborough Hwy on Boulia Rd WA WA: El Questro Station NZ: Trass NC: Sommet Noir, Paagoumène, 11 km NW Koumac QLD: 4.9 km E Georgetown QLD: Kennedy Rd turnoff to Porcupine Gorge QLD: 20.0 km W Gulf Development Rd & Kennedy Hwy NSW: Marramarra National Park NSW: Moonbi Lookout, Moonbi Ranges NSW: Arakoola Nature Reserve QLD: 27 km N Porcupine Ck campground. QLD: Dawson Development Rd, 18 km E Alpha T/off NSW: Warrumbungle National Park, Gould’s circuit WA: 4 km NE of Surveyors Pool, Mitchell Plateau WA: 3.5 km upstream from Bells Gorge, Isdell River. WA: 73 km E Norseman QLD: Kroombit Tops NT: Litchfeild National Park QLD: Townsville GenBank accession numbers RAG1 PDC ND2 AY662627 JQ173721 JQ173722 JQ173723 JQ173725 JX024503 JQ173724 GU459409 JQ173726 JQ173727 JQ173728 JQ173729 — JQ173673 JQ173674 JQ173675 JQ173677 — JQ173676 GU459611 JQ173678 JQ173679 JQ173680 JQ173681 AY369016 JQ173628 JQ173628 JQ173629 JQ173631 JX024362 JQ173630 GU459812 JQ173632 JQ173633 JQ173634 JQ173635 JQ173734 JQ173736 JQ173737 JQ173746 JQ173747 JQ173745 JQ173748 KJ803702 FJ855450 JQ173755 JQ173753 JQ173754 JQ173686 JQ173688 JQ173689 JQ173698 JQ173699 JQ173697 JQ173701 JQ173700 JQ173703 JQ173709 JQ173707 JQ173708 JQ173640 JQ173642 JQ173643 JQ173652 JQ173653 JQ173651 JQ173655 JQ173654 EF681803 JQ173661 JQ173659 JQ173660 Locality Outgroup species Amalosia rhombifer Amalosia rhombifer Nebulifera robusta Oedura tryoni Oedura tryoni Pseudothecadactylus australis Pseudothecadactlus lindneri Rhacodactylus chahoua Rhynchoedura ornate Strophurus assimilis Strophurus ciliaris aberrans Strophurus ciliaris ciliaris Strophurus elderi Strophurus intermedius Strophurus rankini Strophurus spinigerus Strophurus strophurus Woodworthia maculatus Museum number GenBank accession numbers RAG1 PDC ND2 AMS R140413 WA: Roebuck Bay, Broome Bird Observatory JQ173751 JQ173705 JQ173657 AMS R142587 ABTC 3938 (tissue) AMS R152045 AMS R157247 QMJ R57120 AMS R90915 AMS R161238 AMS R155371 AMS R149832 AMS R136023 AMS R147216 AMS R130987 AMS R158434 AMS R140490 AMS R149815 AMS R140536 RAH292 QLD: Lamb Range QLD: near Rathdowney JQ173752 JQ173756 JQ173706 JQ173710 JQ173658 JQ173662 NSW: Moonbi Lookout, Moonbi Ranges NSW: 19.2 km W Tenterfield QLD: Heathlands NT: Liverpool R NC: Dome de Tiebaghi, 14 km NW Koumac NSW: Sturt National Park WA:17.6 W Bonny Vale Railway Station WA: Tanami Hwy NT: Barkley Hwy NSW: 17.9 km N of Coombah Roadhouse NSW: 35 km from Mt Hope WA: Coral Bay WA: Buckland Hill WA: Denham NZ: Titahi Bay JQ173757 JQ173758 HQ288425 AY662626 JQ173759 GU459553 JQ173760 JQ173761 JQ173762 JQ173763 GU459551 JQ173764 JQ173765 JQ173766 GU459449 JQ173711 JQ173712 — — JQ173713 GU459755 JQ173714 JQ173715 JQ173716 JQ173717 GU459753 JQ173718 JQ173719 JQ173720 GU459651 JQ173663 JQ173664 FJ855449 AY369024 JQ173665 GU459954 JQ173666 JQ173667 JQ173668 JQ173669 GU459952 JQ173670 JQ173671 JQ173672 GU459852 Table S2 Details of primers used in this study. Primer pairs Sequence Reference Fragment length PCR conditions ND2.F ND2.R GCC CAT ACC CCG AAA ATS TTG TTA GGG TRG TTA TTT GHG AYA TKC Oliver et al. (2007) Oliver et al. (2007) c. 900 bp 95 °C, 5 min; 40 × (95 °C, 30 s; 55 °C, 30 s; 72 °C, 1 min); 72 °C, 5 min; hold at 15 °C L4437 Asn-trna AAG CTT TCG GGG CCC ATA CC CTA AAA TRT TRC GGG ATC GAG GCC Macey et al. (1997) Read et al. (2001) c. 1200 bp 95 °C, 5 min; 40 × (95 °C, 30 s; 55 °C, 30 s; 72 °C, 1 min); 72 °C, 5 min; hold at 15 °C ND2_grac.F TCC TTC CTA TAC TAA TAA ACC CAG T AGC TTG GTA ATG TGG ATA GGG CT this study c. 1000 bp 95 °C, 5 min; 40 × (95 °C, 30 s; 50 °C, 30 s; 72 °C, 1 min); 72 °C, 5 min; hold at 15 °C PHO.F2 PHO.R1 AGA TGA GCA TGC AGG AGT ATG A TCC ACA TCC ACA GCA AAA AAC TCC T Bauer et al. (2007) Bauer et al. (2007) c. 450 bp 95 °C, 5 min; 40 × (95 °C, 30 s; 50 °C, 30 s; 72 °C, 1 min); 72 °C, 5 min; hold at 15 °C RAG1R13.F RAG1R852.R TCT GAA TGG AAA TTC AAG CTG TT GAG TCT GCA GAA TAA GTG CTT GCA Groth & Barrowclough (1999) c. 1000 bp 95 °C, 5 min; 40 × (95 °C, 30 s; 55 °C, 30 s; 72 °C, 1 min); 72 °C, 5 min; hold at 15 °C RAG1R13.F RAG1R18.R TCT GAA TGG AAA TTC AAG CTG TT GAT GCT GCC TCG GTC GGC CAC CTT T Groth & Barrowclough (1999) Groth & Barrowclough (1999) c. 1000 bp 95 °C, 5 min; 40 × (95 °C, 30 s; 58 °C, 30 s; 72 °C, 1 min); 72 °C, 5 min; hold at 15 °C RAG1R13.F RAG1r.Stroph840 TCT GAA TGG AAA TTC AAG CTG TT AAG TGC TTG CAT GTT GTT TC Groth & Barrowclough (1999) Stuart Nielson1 (unpubl. data) c. 800 bp 95 °C, 5 min; 40 × (95 °C, 30 s; 55 °C, 30 s; 72 °C, 1 min); 72 °C, 5 min; hold at 15 °C RAG1f.Stroph379 RAG1R18.R RAG1f.Stroph681 GTG AGA GGA GAC ATT GAY ACA GAT GCT GCC TCG GTC GGC CAC CTT T TTT GTA CTG AGA TGG ATC TTT TTG CA AAT CTT TTT CAT GAG GCT TTT Stuart Nielson1 (unpubl. data) Stuart Nielson1 (unpubl. data) Stuart Nielson1 (unpubl. data) c. 800 bp 95 °C, 5 min; 40 × (95 °C, 30 s; 55 °C, 30 s; 72 °C, 1 min); 72 °C, 5 min; hold at 15 °C 95 °C, 5 min; 40 × (95 °C, 30 s; 55 °C, 30 s; 72 °C, 1 min); 72 °C, 5 min; hold at 15 °C ND2_grac.R RAG1r.Stroph703 1Stuart Neilson, University of Mississippi. this study Stuart Nielson1 (unpubl. data) c. 800 bp REFERENCES Bauer, A.M., de Silva, A., Greenbaum, E. & Jackman, T. (2007) A new species of day gecko from high elevation in Sri Lanka, with a preliminary phylogeny of Sri Lanka Cnemaspis (Reptilia, Squamata, Gekkonidae). Mitteilungen aus dem Museum für Naturkunde in Berlin, Zoologische Reihe, 83 (S1), 22–32. Groth, J.G. & Barrowclough, G.F. (1999) Basal divergences in birds and the phylogenetic utility of the nuclear RAG-1 gene. Molecular Phylogenetics and Evolution, 12, 115–123. Jennings, W.B., Pianka, E.R. & Donnellan, S. (2003) Systematics of the lizard family Pygopodidae with implications for the diversification of Australian temperate biotas. Systematic Biology, 52, 757–780. Macey, J.R., Larson, A., Ananjeva, N.B., Fang, Z. & Papenfuss, T.J. (1997) Two novel gene orders the role of light-strand replication in the rearrangement of the vertebrate mitochondrial genome. Molecular Biology and Evolution, 14, 91–104. Oliver, P., Hugall, A., Adams, M., Cooper, S.J.B. & Hutchinson, M. (2007) Genetic elucidation of cryptic and ancient diversity in a group of Australian diplodactyline geckos; the Diplodactylus vittatus complex. Molecular Phylogenetics and Evolution, 44, 77–88. Read, K., Keogh, J.S., Scott, I.A.W., Roberts, J.D., Doughty, P. (2001) Molecular Phylogeny of the Australian frog genera Crinia and Geocrinia and allied taxa (Anura: Myobatrachidae). Molecular Phylogenetics and Evolution, 21, 294–308. Table S3 Allozyme frequencies at 38 variable loci for the 11 taxa within the O. marmorata complex, identified as a result of the allozyme PCoAs. Taxon codes follow those in Fig. 4. For polymorphic loci, the frequencies of all but the rarest allozymes are expressed as percentages and shown as superscripts (allowing the frequency of the rare allozyme to be calculated by subtraction from 100%); allozymes with the same frequency are grouped together (e.g. a4, h4 = ah4). Sample sizes for each taxon are shown in parentheses. The following loci were invariant: Enol, Gda, Gdh, Gpd1, Ldh1, Ldh2, Mdh2, Np, Pgam, Pk and Tpi. Locus Central (6) Acon1 Acon2 Acyc Adh1 Adh2 Adh3 Ak Ca Dia Est Fdp Fum Gapd Glo Got1 Got2 Gpd2 Gpi b g a a a c a a b c83, e b d a b b a b a Eastern (8) b b31, d a a d56, c25, a c88, d a a b c88, d b d75, a a b94, a b94, d a b a g44, Gulf 1 (1) Gulf 2 (3) O. gemmata (1) c c c c50, g83, d50, f a e b a a b b e b d b b 50 b ,g a b a e b e83, b b a83, b a b b 50 e , c33, d b d b b 66 d , bc17 a b a a e d a a a b e b d b b c a b a i Western North 1 (5) North 2 (2) North 3 (1) North 4 (2) c g a 50 b , e37, d b a 90 a ,b a90, b b e b d90, a a80, b b 90 b ,e a b a c g a e b75, e a a a75, b b e b d b b c a b a C G A D b a a a b e a50, b d a b b a b a d g a 75 e ,c b a a a b e b d75, b b b c a b75, c a75, b Widespread (39) f59, b99, d e4, ah3, i2, cj1 a88, b a c a a99, c a99, c a84, b 88 c , b10, ae1 b d96, e3, c a b b97, a a92, b b88, a a g27, Burrup (3) b83, a f a a c a a a a c b d a b f83, b c b a Locus Central (6) Gpx Idh1 Idh2 Lap Mdh1 Me1 Me2 Mpi Ndpk1 Ndpk2 PepA1 PepA2 PepB PepD 6Pgd Pgk Pgm1 Pgm2 Sod Sordh b67, d b b c b b 67 c ,d b83, d9, a a b92, a b 92 e ,f d42, c33, b17, a c92, b a83, b a a b b 92 b ,f Eastern (8) Gulf 1 (1) Gulf 2 (3) b b b b c b a h b a b b e c f b a b b b50, c c50, d b b b c b a h b83, d a b b 83 e ,c c f b a b b b d b75, a b c b b62, a c b a b94, c b e c87, b e75, c19, f b a a b b b O. gemmata (1) Western North 1 (5) North 2 (2) North 3 (1) North 4 (2) Widespread (39) Burrup (3) b b a50, c80, b d c50, g c b a 75 e ,f d 75 a ,b b e e c50, b25, e c50, a25, d b a b b b 50 a ,f b d b50, e a b a e b a b b b e50, g c b a b b b f b b b d b a g d75, c a b c e c c b a b b b 75 c ,f b97, c2, a b d53, b46, a c95, b b99, a a 60 21 a , e , b11, d b93, c a96, c b99, a b99, d 88 e , d7, f3, ab1 c67, d21, b11, e c97, a 96 b , c 3, a b63, a 82 b , c15, a b97, d2, c a51, b46, d b89, e9, f b b f c b a e b a b b d c c b a b b b83, d b b b c b a 50 c ,f c a b b50, d e b50, c c b a b a50, b b c d b a b a 50 f , e30, c c80, b a b b60, a e80, g 50 c , e40, f c60, e c90, b a 90 b ,c b90, a b 90 c ,e Table S4 Pairwise genetic distance values among the 11 groups identified by allozyme analysis within the O. marmorata complex. Lower left triangle, number of loci displaying a fixed allozyme difference; upper right triangle, unbiased Nei distance. Taxon Central Eastern Gulf 1 Gulf 2 Central Eastern Gulf 1 Gulf 2 O. gemmata North 1 North 2 North 3 North 4 Western – widespread Western – Burrup — 0* 12 12 11 10 14 12 13 5 0.06 — 12 11 9 10 14 12 13 3 0.32 0.26 — 2 7 7 9 11 9 9 0.33 0.28 0.04 — 8 8 7 11 8 8 0.30 0.25 0.12 0.18 — 6 7 12 6 10 0.28 0.27 0.18 0.22 0.13 — 7 3 7 7 0.36 0.34 0.16 0.18 0.11 0.18 — 8 6 11 0.31 0.30 0.23 0.27 0.24 0.12 0.18 — 11 9 10 8 13 14 14 13 15 13 O. gemmata North 1 North 2 North 3 North 4 0.35 0.33 0.18 0.21 0.12 0.18 0.14 0.26 — 11 16 Western Widespread 0.17 0.14 0.24 0.28 0.25 0.27 0.31 0.26 0.31 — 3 Burrup 0.28 0.23 0.31 0.36 0.32 0.36 0.36 0.31 0.38 0.10 — *Although these taxa did not display any fixed differences, they did exhibit major differences in allele frequency (p > 80%) at two allozyme loci plus moderate differences (p > 50%) at a further three loci (Table S3).