HCV
advertisement

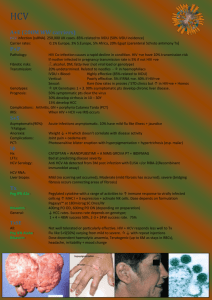
GCAT-SEEKquence The Genome Consortium for Active Teaching NextGen Sequencing Group NextGen Sequencing Request Form Complete fields below, save file with your last name at the beginning of the filename (e.g. newman-GCAT-SEEK Sequence request form.pdf) and email to Vincent Buonaccorsi <BUONACCORSI@juniata.edu> A. Contact Information 1. Name: Dennis Revie 2. Department: Biology 3. Institution: California Lutheran University 4. Phone Number: 805-493-3380 5. Email Address: revie@clunet.edu B. Project Information 1. Title: Analysis of the response of monocytes to Hepatitis C Virus (HCV) 2. Category: RNA-seq/transcriptomes 3. Total amount of sequence requested: 1 to 3 lanes 4. Preferred technology: Illumina 5. Do you have funds for a partial run next Spring? No C. Describe the background, hypotheses and specific aims (500 words max) HCV is a positive strand RNA virus that infects humans. We previously developed a system for the isolation of HCV and its replication in vitro (Revie et al, 2005). The system uses serum from infected patients to infect macrophages. Recently, we have extended the system to the use of monocytic cell lines for culturing HCV. Macrophages derive from monocytes and they can differentiate into macrophages by the addition of PMA (phorbal myristate acetate). These cell lines have the advantage of being available to all researchers. We found that HCV can infect a U-937 monocytic cell line while it does not infect a monocytic THP-1 DC cell line. We are interested in examining the response of the monocytes to infection by HCV. First, we want to compare the two monocytic cell lines to determine the differences in gene expression between the two cell lines. In particular, we are interested in differences between cell surface proteins that might affect virus binding and entry or in genes involved in the cellular response to viral infections. Second, we will compare gene expression in cells that are infected by HCV with those that are not. This will let us determine which genes are up or down regulated in response to the infection. These studies may help explain why some cell types can be infected by HCV while others cannot. D. Describe the methods [sample prep, calculation of amount of sequence required, analysis plan] We will analyze uninfected U-937, HCV-infected U-937 at five time points post-infection (0, 1, 4, 16 and 64 hours), uninfected THP-1, and infected THP-1 at three time points post-infection (0, 1, and 4 hours). This will give us 10 samples. At the specified times, RNA will be isolated for the analysis. We also plan to perform the experiments in triplicate to provide better statistical comparisons between the samples as well as to check for reproducibility. As one lane of Illumina can give 37 GBp or more of 75 bp sequence, we can barcode the samples to reduce the number of lanes needed for the analyses. If we put 10 barcoded samples/lane, 1 lane would provide one full set of data, while 3 lanes would give us triplicate results (Wang et al., 2011). Comparisons of genes expressed in the two uninfected cell lines will provide candidate genes that may be important for infection by HCV. Comparisons of infected and uninfected cell lines will provide gene candidates that may be involved in combating the virus. As the genes expressed will change with the time of infection, a time course should show differences for genes expressed early and later in infection. For example, the virus must enter the cell, disassemble, replicate, reassemble, and then exit the cell. Each step in the process may activate cellular genes that either help or hinder the process. Gene candidates can then be compared to ones identified by other researchers for hepatoma cells (e.g., Woodhouse et al., 2010). E. Describe the role and number of undergraduates involved in the project, and how they would benefit. All aspects of the project will be performed by undergraduates. The cell culturing and RNA purification will be performed by undergraduate research students. The data analysis will be done by undergraduate researcher students and also by students in a Genomics course (Biology 427). Different students or groups of students will test different hypotheses, such as which membrane receptors differ between U-937 cells and THP-1 cells, which antiviral pathways are activated by the infection process in the two cell types, etc. I typically have around 6 undergraduates per year work on research projects, and the Genomics course has about 10 students per year. F. I agree to administer the GCAT-SEEK pre- and post-activity assessment test for students and to complete the faculty post-utilization survey. __x__ yes, ____ no G. Describe any other broader impact or intellectual merit considerations. HCV is estimated to infect 170 million individuals in the world, and it is estimated that about 3 million are infected in the US. HCV can cause liver diseases as well as extrahepatic diseases such as B cell lymphomas. The major site of HCV production in individuals is the liver, but extrahepatic replication of HCV also occurs (Revie and Salahuddin, 2011). To date, only one RNA-seq study has been published that has investigated the response of cells to HCV in vitro (Woodhouse et al., 2010), and it used a synthetic virus to infect hepatoma cells. Therefore, this study will provide the first RNA-seq data for infection of non-liver cells, the first to analyze cellular response over time, and the first to use HCV from patient serum instead of a synthetic virus. H. References Revie D, Braich RS, Bayles D, Chelyapov N, Khan R, Geer C, Reisman R, Kelley AS, Prichard JG, Salahuddin SZ (2005). Transmission of human hepatitis C virus from patients in secondary cells for long term culture. Virol Journal 2005, 2:37. Revie D, Salahuddin SZ (2011). Human cell types important for Hepatitis C Virus replication in vivo and in vitro. Old assertions and current evidence. Virology Journal 8:346 Wang Y, Ghaffari N, Johnson CD, Braga-Neto UM, Wang H, Chen R, Zhou H. Evaluation of the coverage and depth of transcriptome by RNA-Seq in chickens. BMC Bioinformatics. 2011 Oct 18;12 Suppl 10:S5. Woodhouse SD, Narayan R, Latham S, Lee S, Antrobus R, Gangadharan B, Luo S, Schroth GP, Klenerman P, Zitzmann N. (2010). Transcriptome Sequencing, Microarray, and Proteomic Analyses Reveal Cellular and Metabolic Impact of Hepatitis C Virus Infection In Vitro. Hepatology 52, 443-453.