Table S1. - BioMed Central
advertisement

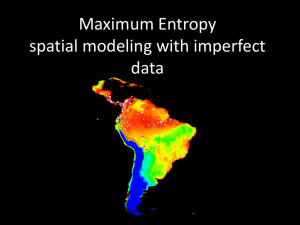
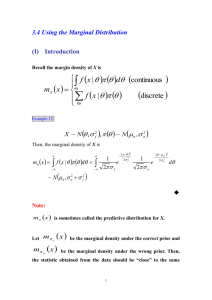
Additional file Tables Table S1. Loci used in this study Table S2. Summary statistics and neutrality tests of populations at species boundaries Table S3. Other priors tested in IMa2 Table S4. Model evaluation for ecological niche modeling Figures Figure S1. Locations of occurrence data used for ecological niche modeling Figure S2. Projection of current and past distribution of R. palmatus and R. grayanus using the Maxent Tables Table S1. Loci used in this study Loci used and summary statistics at species boundaries and contact zone (Table 1). The putative gene functions are annotated based on the database of Arabidopsis thaliana reference proteins (min. threshold e-value was less than 10-20). bp: total length of sequence used for analysis; n(total) the total number of phased sequences; S: the total number of segregating sites; fixed S: the number of segregating sites between species that are fixed within a species; FST : population differentiation. COP1- and GSTF-homolog gene sequences included non-coding regions. Asterisk indicates chloroplast DNA region. Contact Zones putative gene function bp n S number of haplotype θ(total) π(total) CO 305 154 6 9 0.0041 0.0067 COP1 276 158 5 6 0.0032 0.0046 CCNB1 hypothetical protein ETR 118 158 4 4 0.0060 0.0122 482 160 9 8 0.0035 0.0070 258 328 154 158 3 7 4 4 0.0021 0.0037 0.0026 0.0077 349 639 158 158 7 9 10 7 0.0036 0.0025 0.0054 0.0042 PHYA 416 362 160 158 10 7 5 6 0.0042 0.0034 0.0107 0.0077 PAL4 603 160 15 13 0.0044 0.0054 LDOX 393 154 4 4 0.0018 0.0035 PHR2 518 158 9 11 0.0038 0.0045 EXPA 237 160 4 5 0.0030 0.0047 XET 248 160 5 5 0.0036 0.0067 hypothetical protein 276 154 2 3 0.0013 0.0034 IAA 351 160 3 4 0.0015 0.0026 TrnH_psbA* 238 80 3 2 0.0030 0.0053 F3H F3GT GI GSTF total 6302 112 Table S2. Summary statistics and neutrality tests of populations at species boundaries Population summary statistics for the populations at the species boundaries (Table 1). The parameters shown are the number of haplotype studied: n; the average number of segregating sites: S; nucleotide diversity (θ and π) for total, synonymous (s), non-synonymous (a), and non-cording. Tajima’s D and Fu & Li D were also calculated and simulated. Mean values and lower and higher 95% intervals were simulated with 1000 coalescent simulations and compared with the observed value to test its significance. +p<0.10, *p < 0.05, **p < 0.01. Table S3. Other priors tested in IMa2 Other priors tested in this study and highest probability density (HPD) with 95% HPD in parentheses, in isolation with migration models of two species populations (pop1 and pop2): divergence time in million years ago (t) of two focal populations, population size in thousands in population1 (θ1), population 2 (θ2), ancestral population (θA), and migration rate from population 2 to 1 (2NM1<2) and from population 1 to 2 (2NM1>2). Migration rates (2NM) were tested by likelihood ratio tests (Nielsen and Wakeley, 2001); *p < 0.05, **p < 0.01, ***p < 0.001. We used two migration priors for independent runs; a uniform distribution on [0,1] for migration rate, and an exponential distribution with mean m* = 0.05. model pop1, 2 run1 pYK, pEB prior θ 10 posterior m t hn 1 4 20 1 5 40 run2 10 run3 10 0.05 5 40 4 2 2 20 10 1 5 40 5 0.05# 4 40 run4 # gAM, gYK run5 run6 run7 pYK, gAM run8 run9 run10 pYK, gYK t θ1 θ2 θA 2NM1<2 2NM1>2 0.14 (0.03-3.63) 18.8 (9.3-32.0) 28.8 (16.1-45.7) 54.3 (0-255.2) 0.020 (0-0.216) 0.223 (0-0.362) 0.15 (0.03-4.54) 18.4 (9.3-32.0) 28.9 (13.6-45.7) 54.8 (0-288.4) 0.031 (0-0.215) 0.227 (0-0.363) 0.11 (0.05-0.21) 17.5 (8.4-31.1) 27.5 (14.8-46.1) 59.8 (34.8-96.1) 0.000 (0-0.035) 0.000 (0-0.073) 0.016 (0.003-0.13) 5.4 (1.3-14.1) 11.1 (2.9-27.6) 22.4 (7.9-42.6) 0.034 (0-0.235) 0.054 (0-0.477) 0.015 (0.006-0.07) 4.8 (1.5-14.0) 10.2 (2.7-26.5) 22.3 (8.9-40.2) 0.010 (0-0.104) 0.007 (0-0.212) 0.015 (0.006-0.065) 4.8 (1.5-13.5) 9.8 (2.7-26.5) 22.3 (9.4-40.2) 0.000 (0-0.013) 0.000 (0-0.025) 1 5 40 10 0.05# 5 40 10 1 5 40 1.15 (0.42-4.59) 42.2 (26.7-62.9) 12.9 (6.0-25.0) 20.9 (0-348.4) 0.000 (0-0.106) 0.000 (0-0.079) 0.99 (0.44-1.67) 45.1 (29.0-66.4) 15.8 (7.7-27.9) 34.2 (0-123.1) 0.000 (0-0.041) 0.000 (0-0.019) 1.00 (0.76-4.74) 12.7 (6.2-23.4) 13.9 (6.8-25.8) 12.1 (0-528.7) 0.072*** (0.028-0.131) 0.015** (0.001-0.070) 10 0.05# 5 40 1.07 (0.27-2.22) 17.5 (8.6-29.9) 16.9 (8.6-29.3) 0.9 (0 - 170.3) 0.022*** (0.005-0.057) 0.005** (0.0003-0.026) 10 Table S4. Model evaluation for ecological niche modeling Averaged TSS, AUC and Kappa values for model evaluation. The values were averaged from 10 runs of each modeling technique for each of 5 sets of pseudo-absence datasets. model GLM GAM MARS CTA RF MAXENT R. palmatus TSS AUC KAPPA 0.850 0.846 0.847 0.818 0.854 0.839 0.960 0.960 0.959 0.926 0.971 0.960 0.862 0.826 0.875 0.740 0.960 0.946 R. grayanus TSS AUC KAPPA 0.937 0.959 0.961 0.940 0.998 0.997 0.969 0.979 0.987 0.970 0.999 0.997 0.859 0.857 0.855 0.837 0.865 0.854 Figures Figure S1. Locations of occurrence data used for ecological niche modeling. Red and green circles represent occurrence data for R. palmatus and R. grayanus, respectively. Occurrence data were obtained from GBIF (http://www.gbif.org) and also included the sampling sites for phylogeographic analysis in this study. 0 25 50 75 100 Figure S2. Projection of current (a, b) and past (c, d) distribution of R. palmatus and R. grayanus using the Maxent. Maxent is a modeling technique that use only presence data. A dozen of runs were cross-validated with random seeds and averaged for each species projection.