Regulation of Gene Expression
advertisement

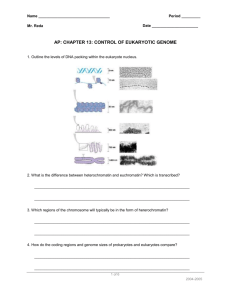
Independent assortment Sex Crossing over Alternative splicing Mutations Sending it to different environments Acetylation of histone tails – lose and the gene is expressed Methylation of the gene – controls its transcription Random fertilization Regulation of Gene Expression: Operons: o A gene with a promoter and an operator o Operator – works with something else to control the genes First 2 Operons: o Tryptophan (amino acid) operon – default setting is turned on, but when there’s tryptophan, the bacteria turn it off sometimes. o Lac operon Upstream of a DNA strand means the RNA polymerase is going the other way Operator also controls the RNA polymerase whether it’ll run through the upstream or not Tryptophan Operon: TrypR is a regulatory gene Tryptophan makes tryptophan. The repressor protein is inactive. When tryptophan binds to the repressor protein, the repressor protein is active. Then, it binds with the operator. Then it blocks RNA polymerase. Regulatory gene – the Corepressor Lac Operon in E.coli: The default is turned off (the regulatory protein is active) The inducer binds with the repressor, which makes it inactive No milk, no lactase Have milk, have lactose Allolactose is the inducer, induces the lactase by binding to the repressor 18.5 diagram cAMP: Cyclic AMP (gay) – running out of sugar/last phosphate CAP protein is inactive, cAMP will bind and it’ll be activated Active CAP activates the RNA polymerase/activates the gene Get rid of the repressor and activator must bound to the promoter in order for the gene to be expressed We have lactase, no lactose, no glucose Prokaryotes regulates expression at the transcription level Eukaryotes regulates expression at transcription, RNA processing and translation. Small interfering RNAs or micro RNAs – interferes or stops proteins by complementarying the piece of protein, so that the protein won’t go to the ribosome Histone modifcations – The more acetylation, the more expression you get Acetylation and methylation can control the gene expression 18.6: Demethylation and acetylation increases the gene expression Transcription – primary transcript will control the expression RNA processing – Transport to cytoplasm – Translation – Secondary, tertiary, and quaternary structure – Destination of the cell – Attacking/Degrading the protein happens after expressing the gene Epigenetic Inheritance: Something outside of your gene that can be inherited Inheritance of traits transmitted by mechanisms not directly involving the nucleotide sequence If the thing that controls the gene isn’t coming from the cell’s genome 18.8 Diagram: Eukaryotic gene and its transcript Termination region is in a eukaryotic gene – area where termination stops Enhancer (increase some property) – they act like promoters (attract RNA polymerase) If those enhancers move or bend, then it’ll be next to promoters, which will attract even more RNA polymerase (more transcription) Activators are chemicals that binds to the enhancer region and causes it to bend to the promoter 18.9 Diagram: Distal control element – far away from the operon DNA bending protein – bends the DNA Activators are now over the promoter RNA polymerases are attracted One gene can make more than 1 different genes 18.10 Diagram: Albumin gene is in livers o Activators are coming from the liver o Binds to the enhancer making the albumin gene expressed, crystalline gene isn’t expressed Crystalline gene is in lens or eyes o Activators bind to the enhancer making the crystallin gene expressed, but albumin gene isn’t expressed mRNA degradation Initiation of Translation – Protein Processing and Degrading – 18.12 Diagram: Ubiquitin is the kiss of death Once the ubiquitin binds to the protein, the protein will enter the proteasome The protein will be destroyed/destroyed microRNAs Small single-stranded RNA molecules RNA interference or small interfering RNAs Ds Cell Differentiation – the process by which cells become specialized in structure and function is based on the activators and the transcription factors Morphogenesis – creation of form Cytoplasmic determinants – maternal substances in the egg that influence the course of early development Cytoplasmic determinants give morphogenesis Induction makes the something same, gives determination 18.16 Diagram: Embryonic stem cell is not determined and not differentiated Adult stem cell is determines, but not differentiated (Myoblast) Pattern formation – Cytoplasmic determinants and inductive signals both contribute to the development of a spatial organization in which the tissues and organs of an organism are all in their characteristic places Positional information – the molecular cues that control pattern formation Homeotic genes – control whole set of patterns of genes Embryonic lethals – mutations with phenotypes causing death at the embryonic or larval stage Maternal effect gene – genes from the mom that will affect the youngster Egg-polarity genes – knowing the top and the end Bicoid – two-tailed Morphogens – any substances that changes the shape (which is the head which is the tail) Cancer – disruption of the cell cycle Oncogenes – genes that cause cancer (problems with certain genes lead to cancer) Proto-oncogenes – code for proteins that stimulate normal cell growth and division Tumor-suppressor genes – inhibit cell division, makes proteins that suppress the cell cycle MPF and CDK (mitosis ch. 14) 18.20 Diagram: Translocation or transposition (epistasis) can cause oncogene Lose control of its expression due to the moving of its locus 18.21 Diagram: Cell cycle-stimulating pathway Cell cycle-inhibiting pathway – UV light comes in and damages the genome Point mutation Ras gene – a gene that’s related to cancer found in rats (uncogene) P53 gene – gene found in humans (uncogenes) One cancer can lead to many other cancers